system.file(package = "ROracle")[1] "/home/haf/einarhj/R/x86_64-pc-linux-gnu-library/4.3/ROracle"system.file(package = "mar")[1] "/home/haf/einarhj/R/x86_64-pc-linux-gnu-library/4.3/mar"To access the MFRI database you need:
Testing if you have {ROracle} and {mar} installed:
[1] "/home/haf/einarhj/R/x86_64-pc-linux-gnu-library/4.3/ROracle"[1] "/home/haf/einarhj/R/x86_64-pc-linux-gnu-library/4.3/mar"Ideally these should be installed by the IT-department when they set up your computer, including R and RStudio.
If you do not have ROracle do:
If you do not have mar do:
The mar_tables-function gives us some informations about what is in the database:
Rows: ??
Columns: 6
Database: OraConnection
$ owner <chr> "sys", "sys", "sys", "sys", "sys", "sys", "sys", "sys"…
$ table_name <chr> "dual", "system_privilege_map", "table_privilege_map",…
$ comments <chr> NA, "Description table for privilege type codes. Maps…
$ tablespace_name <chr> "SYSTEM", "SYSTEM", "SYSTEM", "SYSTEM", "SYSTEM", NA, …
$ num_rows <dbl> 1, 257, 26, 4, 337, NA, NA, 1, 239, 457, 0, NA, NA, NA…
$ last_analyzed <dttm> 2023-06-23 22:00:59, 2021-04-30 22:00:37, 2021-04-26 …Here we have among other things:
Let’s see the schemas we have and how many tables in each schema:
info |>
count(owner, name = "n.tables") |>
arrange(owner) |>
collect() |>
knitr::kable(caption = "List of owners and the number of tables")| owner | n.tables |
|---|---|
| adb | 15 |
| afli | 19 |
| agf | 3 |
| asfis | 1 |
| ask | 9 |
| ata | 28 |
| bathymetry | 1 |
| biota | 35 |
| botndyr | 27 |
| channel | 35 |
| ctxsys | 5 |
| erfdafraedi | 8 |
| fiskar | 4 |
| fiskifelagid | 2 |
| fiskmerki | 21 |
| flokkun | 14 |
| fuglar | 8 |
| gear | 18 |
| hafmynd | 26 |
| hafro | 10 |
| hafvog | 3 |
| hvalir | 2 |
| hydro | 40 |
| kvoti | 19 |
| mdsys | 57 |
| merki | 24 |
| nyting | 3 |
| ops\(anika | 7| |ops\)bthe | 46 |
| ops\(einarhj | 51| |ops\)ella | 3 |
| ops\(krik | 2| |ops\)mdan | 1 |
| ops\(pamela | 8| |ops\)rafn | 3 |
| ops\(sigfus | 4| |ops\)will | 9 |
| orri | 9 |
| pame | 1 |
| phyto | 13 |
| selir | 14 |
| siritar | 5 |
| stk | 13 |
| svifthorungar | 17 |
| syni | 15 |
| sys | 45 |
| system | 41 |
| taxon | 7 |
| telogger | 2 |
| thorungar | 15 |
| ufs | 2 |
| veidibok | 10 |
| vessel | 18 |
| worms | 1 |
| xdb | 6 |
Let’s check what tables are in phyto:
info |>
filter(owner == "phyto") |>
select(table_name, comments) |>
arrange(table_name) |>
collect()# A tibble: 13 × 2
table_name comments
<chr> <chr>
1 dypi "Í töflunni eru upplýsingar ýmissa umhverfisþátta, sem hafa veri…
2 hafur "Í töflunni eru upplýsingar um hvaða þörungategundir hafa fundis…
3 observation <NA>
4 prod "Tafla eftir eyðslu á dálkunum year og turner"
5 prod_old "Gögn eftir eyðslu á year og the_date > '83 gögnum"
6 stadur "Í töflunni eru upplýsingar um staðarnöfn þar sem mælingar hafa\…
7 station <NA>
8 station_old <NA>
9 stodvar "Í töflunni eru upplýsingar um stað og stund þar sem sýnataka fe…
10 talning "Í töflunni eru upplýsingar um fjölda einstaklinga af tiltekinni…
11 tegundir "Í töflunni er ein færsla fyrir hverja þörungategund. Ýmiss kona…
12 tmp_obs <NA>
13 underway <NA> Because the construct of tables within a database can be quite complex, a view is often created that may combine different tables. The intent is to make it easier for the downstream user to operate on the data. Let’s check what views are in veidibok:
| owner | view_name |
|---|---|
| veidibok | fisktegundir_v |
| veidibok | notendur_v |
| veidibok | vbs_vf_dagveidi_v |
| veidibok | vbs_vf_fjoldi_kyn_heiti_v |
| veidibok | vbs_vf_fjoldi_kyn_v |
| veidibok | vbs_vf_fjoldi_tegund_heiti_v |
| veidibok | vbs_vf_fjoldi_tegund_v |
| veidibok | vbs_vf_lengdardreif_heiti_v |
| veidibok | vbs_vf_lengdardreifing_v |
| veidibok | vbs_vf_stadur_veidi_heiti_v |
| veidibok | vbs_vf_stadur_veidi_v |
| veidibok | vbs_vf_thyngdardreif_heiti_v |
| veidibok | vbs_vf_thyngdardreifing_v |
| veidibok | vbs_vf_thyngdardreif_kyn_v |
| veidibok | vbs_vf_thyngdardr_kyn_heiti_v |
| veidibok | vbs_vf_thyngdartolur_v |
| veidibok | vbs_vf_thyngd_heiti_v |
| veidibok | vbs_vf_vikuveidi_heiti_v |
| veidibok | vbs_vf_vikuveidi_v |
| veidibok | vbs_vs_dagveidi_v |
| veidibok | vbs_vs_fjoldi_kyn_heiti_v |
| veidibok | vbs_vs_fjoldi_kyn_v |
| veidibok | vbs_vs_fjoldi_tegund_heiti_v |
| veidibok | vbs_vs_fjoldi_tegund_v |
| veidibok | vbs_vs_lengdardreif_heiti_v |
| veidibok | vbs_vs_lengdardreifing_v |
| veidibok | vbs_vs_stadur_veidi_heiti_v |
| veidibok | vbs_vs_stadur_veidi_v |
| veidibok | vbs_vs_thyngdardreif_heiti_v |
| veidibok | vbs_vs_thyngdardreifing_v |
| veidibok | vbs_vs_thyngdardreif_kyn_v |
| veidibok | vbs_vs_thyngdardr_kyn_heiti_v |
| veidibok | vbs_vs_thyngdartolur_v |
| veidibok | vbs_vs_thyngd_heiti_v |
| veidibok | vbs_vs_vikuveidi_heiti_v |
| veidibok | vbs_vs_vikuveidi_v |
| veidibok | veidibok_v |
| veidibok | veidi_samtals_v |
| veidibok | veidi_v |
Here we use the mar_fields-function. Lets pick the table “phyto.hafur”
# Source: SQL [?? x 4]
# Database: OraConnection
owner table_name column_name comments
<chr> <chr> <chr> <chr>
1 phyto talning snt <NA>
2 phyto talning snn <NA>
3 phyto talning sbt <NA>
4 phyto talning sbn <NA>
5 phyto talning s_id Númer sem tengir færsluna við ákveðna stöð, lyk…
6 phyto talning dypi <NA>
7 phyto talning skst Skammstöfun tegundarinnar, lykill í TEGUNDIR.
8 phyto talning fjoldi Fjöldi fruma á líter.
9 phyto talning dags_taln Dagsetning talningar.
10 phyto talning rummal Rúmmál talningarhólks í ml.
# ℹ more rowsHere we use the function tbl_mar:
# Source: SQL [?? x 6]
# Database: OraConnection
id taudkenni nr dagstimi dypi hitastig
<dbl> <chr> <dbl> <dttm> <dbl> <dbl>
1 8299166 1C1426 6362 2004-06-13 17:30:00 171. 3.34
2 8299167 1C1426 6363 2004-06-13 17:40:00 168. 3.34
3 6763843 2C0669 43559 2005-02-11 03:00:00 212. 6.35
4 8299168 1C1426 6364 2004-06-13 17:50:00 169. 3.34
5 8299169 1C1426 6365 2004-06-13 18:00:00 168. 3.34
6 6763846 2C0669 43560 2005-02-11 03:10:00 214. 6.35
7 8299170 1C1426 6366 2004-06-13 18:10:00 168. 3.34
8 8299171 1C1426 6367 2004-06-13 18:20:00 168. 3.38
9 6763849 2C0669 43561 2005-02-11 03:20:00 213. 6.35
10 8299172 1C1426 6368 2004-06-13 18:30:00 168. 3.38
# ℹ more rowsmar_tables-, mar_views-, mar_fields, tbl_mar-functions to explore something of your interestThe basic function when we access a database table is tbl. Let’s use to explain what is happening under the hood, here taking a peek at a simple “lookup-table”:
Rows: ??
Columns: 5
Database: OraConnection
$ USAGE_CATEGORY_NO <int> 231, 154, 6299, 6283, 6252, 6237, 6289, 46, 50, 55, …
$ CODE <dbl> 116, 115, 129, 118, NA, 117, 132, 201, 202, 203, 204…
$ NAME <chr> "A - FARÞEGASKIP", "A - FLUTNINGASKIP", "A - FLUTNIN…
$ ENG_NAME <chr> "PASSENGER SHIP", "CARGO SHIP", "Other cargo ship", …
$ CATEGORY <chr> "OtherShip", "OtherShip", "OtherShip", "OtherShip", …Take note here that:
So we have kind of a table but not quite the tables we have so far worked with in R. Check out this e.g.:
[1] "tbl_OraConnection" "tbl_dbi" "tbl_sql"
[4] "tbl_lazy" "tbl" <SQL>
SELECT *
FROM "VESSEL"."USAGE_CATEGORY"Effectively what we have is an SQL-query. Let’s add a filter:
q2 <-
q |>
filter(CATEGORY == "FishingShip") |>
select(name = NAME, english = ENG_NAME, cat = CATEGORY)
q2 |> glimpse()Rows: ??
Columns: 3
Database: OraConnection
$ name <chr> "FISKISKIP", "FRÍSTUNDAFISKISKIP", "HVALVEIÐISKIP", "NÓTAVEIÐI…
$ english <chr> "FISHING VESSEL", NA, NA, NA, "Research Vessel", "TRAWLER", NA
$ cat <chr> "FishingShip", "FishingShip", "FishingShip", "FishingShip", "F…<SQL>
SELECT "NAME" AS "name", "ENG_NAME" AS "english", "CATEGORY" AS "cat"
FROM "VESSEL"."USAGE_CATEGORY"
WHERE ("CATEGORY" = 'FishingShip')So now we have more SQL-code. The big news here is:
Note though that not every code in R is translated to SQL
Normally we would not access a table by the tbl-function, user rather the tbl_mar-function, e.g.:
Rows: ??
Columns: 5
Database: OraConnection
$ usage_category_no <int> 231, 154, 6299, 6283, 6252, 6237, 6289, 46, 50, 55, …
$ code <dbl> 116, 115, 129, 118, NA, 117, 132, 201, 202, 203, 204…
$ name <chr> "A - FARÞEGASKIP", "A - FLUTNINGASKIP", "A - FLUTNIN…
$ eng_name <chr> "PASSENGER SHIP", "CARGO SHIP", "Other cargo ship", …
$ category <chr> "OtherShip", "OtherShip", "OtherShip", "OtherShip", …Sometimes there is a shortcut available, like:
Rows: ??
Columns: 37
Database: OraConnection
$ fiskur_id <dbl> 56255, 56256, 56257, 56258, 56259, 56260, 56261, 56262, 5…
$ synis_id <dbl> 236952, 236952, 236952, 236952, 236952, 236952, 236952, 2…
$ tTegund <dbl> 23, 23, 23, 23, 23, 23, 23, 23, 23, 23, 23, 23, 23, 23, 2…
$ tLengd <dbl> 39, 39, 38, 32, 42, 40, 36, 42, 49, 47, 31, 50, 35, 40, 3…
$ tThyngd <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, …
$ tKyn <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ tKynthroski <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ audkenni <chr> "IH", "IH", "IH", "IH", "IH", "IH", "IH", "IH", "IH", "IH…
$ numer <dbl> 1122, 1775, 1634, 1316, 1565, 1137, 1656, 1830, 1922, 110…
$ leidangur <chr> "B3-73", "B3-73", "B3-73", "B3-73", "B3-73", "B3-73", "B3…
$ stod <dbl> 1424, 1424, 1424, 1424, 1424, 1424, 1424, 1424, 1424, 142…
$ tDags <dttm> 1973-03-07, 1973-03-07, 1973-03-07, 1973-03-07, 1973-03-…
$ tAr <dbl> 1973, 1973, 1973, 1973, 1973, 1973, 1973, 1973, 1973, 197…
$ tReitur <dbl> 422, 422, 422, 422, 422, 422, 422, 422, 422, 422, 422, 42…
$ tSmareitur <dbl> 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, …
$ tLon <dbl> -22.95, -22.95, -22.95, -22.95, -22.95, -22.95, -22.95, -…
$ tLat <dbl> 64, 64, 64, 64, 64, 64, 64, 64, 64, 64, 64, 64, 64, 64, 6…
$ tDypi <dbl> 95, 95, 95, 95, 95, 95, 95, 95, 95, 95, 95, 95, 95, 95, 9…
$ tVeidarfaeri <dbl> 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, 7, …
$ rid <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ rDags <dttm> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, …
$ rAr <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ rManudur <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ rLon <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ rLat <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ rReitur <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ rSmareitur <int> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ rLandshluti <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ rHafsvaedi <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ rVeidarfaeri <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ rDypi <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ rTegund <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ rLengd <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ rKyn <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ rKynthroski <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ rAge <dbl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
$ dst_id <chr> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…<SQL>
SELECT
"FISKAR"."ID" AS "fiskur_id",
"FISKAR"."SYNIS_ID" AS "synis_id",
"TEGUND" AS "tTegund",
"LENGD" AS "tLengd",
"THYNGD" AS "tThyngd",
"KYN" AS "tKyn",
"KYNTHROSKI" AS "tKynthroski",
"AUDKENNI" AS "audkenni",
"NUMER" AS "numer",
"leidangur",
"stod",
"tDags",
"tAr",
"tReitur",
"tSmareitur",
"tLon",
"tLat",
"tDypi",
"tVeidarfaeri",
"rid",
"rDags",
"rAr",
"rManudur",
"rLon",
"rLat",
"rReitur",
"rSmareitur",
"rLandshluti",
"rHafsvaedi",
"rVeidarfaeri",
"rDypi",
"rTegund",
"rLengd",
"rKyn",
"rKynthroski",
"rAge",
"TAUDKENNI" AS "dst_id"
FROM "FISKMERKI"."FISKAR"
LEFT JOIN "FISKMERKI"."MERKI"
ON ("FISKAR"."ID" = "MERKI"."FISKUR_ID")
LEFT JOIN (
SELECT
"SYNIS_ID" AS "synis_id",
"LEIDANGUR" AS "leidangur",
"STOD_V"."STOD_ID" AS "stod",
"DAGS" AS "tDags",
"AR" AS "tAr",
"REITUR" AS "tReitur",
"SMAREITUR" AS "tSmareitur",
"KASTAD_LENGD" AS "tLon",
"KASTAD_BREIDD" AS "tLat",
"BOTNDYPI_KASTAD" AS "tDypi",
"VEIDARFAERI" AS "tVeidarfaeri"
FROM "CHANNEL"."STOD_V"
LEFT JOIN "CHANNEL"."SYNI_V"
ON ("STOD_V"."STOD_ID" = "SYNI_V"."STOD_ID")
) "...3"
ON ("FISKAR"."SYNIS_ID" = "...3"."synis_id")
LEFT JOIN (
SELECT
"rid",
"tid",
"rDags",
"rAr",
"rManudur",
-geoconvert1("rLon" * 100.0) AS "rLon",
geoconvert1("rLat" * 100.0) AS "rLat",
"rReitur",
"rSmareitur",
"rLandshluti",
"rHafsvaedi",
"rVeidarfaeri",
"rDypi",
"rTegund",
"rLengd",
"rKyn",
"rKynthroski",
"rAge"
FROM (
SELECT
"ID" AS "rid",
"MERKI_ID" AS "tid",
"DAGS_FUNDID" AS "rDags",
"AR" AS "rAr",
"MANUDUR" AS "rManudur",
"VLENGD" AS "rLon",
"NBREIDD" AS "rLat",
"REITUR" AS "rReitur",
"SMAREITUR" AS "rSmareitur",
"LANDSHLUTI_ID" AS "rLandshluti",
"HAFSVAEDI_ID" AS "rHafsvaedi",
"VEIDARFAERI_ID" AS "rVeidarfaeri",
"DYPI" AS "rDypi",
"TEGUND" AS "rTegund",
"LENGD" AS "rLengd",
"KYN" AS "rKyn",
"KYNTHROSKI" AS "rKynthroski",
"ALDUR" AS "rAge"
FROM "FISKMERKI"."ENDURHEIMTUR"
) "q01"
) "...4"
ON ("MERKI"."ID" = "...4"."tid")
LEFT JOIN "FISKMERKI"."RAFAUDKENNI"
ON ("MERKI"."ID" = "RAFAUDKENNI"."MERKI_ID")In both cases take note that the variable names now are all in small-caps (they are originally all-caps in Oracle).
Rows: ??
Columns: 6
Database: OraConnection
$ id <dbl> 8641662, 8641663, 8641664, 8641665, 8641666, 8641667, 864166…
$ taudkenni <chr> "1C0407", "1C0407", "1C0407", "1C0407", "1C0407", "1C0407", …
$ nr <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 1…
$ dagstimi <dttm> 2003-04-15 00:00:00, 2003-04-15 00:10:00, 2003-04-15 00:20:…
$ dypi <dbl> 15.44, 15.44, 15.67, 15.44, 15.67, 15.67, 15.67, 15.67, 15.9…
$ hitastig <dbl> 7.59, 7.59, 7.59, 7.59, 7.59, 7.59, 7.59, 7.59, 7.59, 7.59, …q |>
collect() |>
ggplot(aes(dagstimi, -dypi)) +
geom_point(size = 0.1) +
labs(title = "Depth profile from one electronically tagged fish")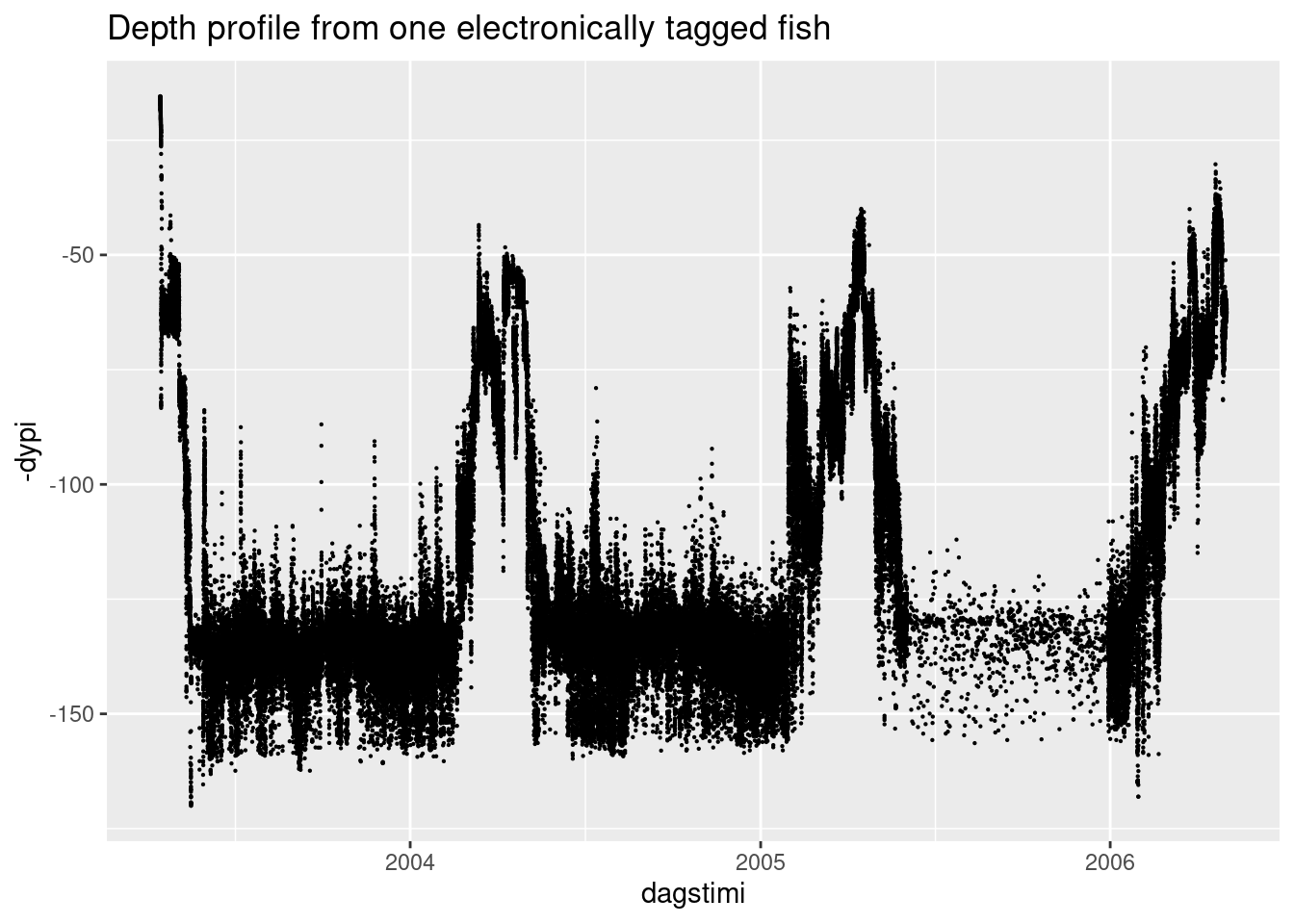
q <-
tbl_mar(con, "hydro.v_sonda") |>
filter(between(latitude, 66, 67),
between(longitude, -19, -18))
q |> glimpse() Rows: ??
Columns: 9
Database: OraConnection
$ l_id <chr> "B6-89", "B6-89", "B6-89", "B6-89", "B6-89", "B6-89", "B6-…
$ id <int> 479, 479, 479, 479, 479, 479, 479, 479, 479, 479, 479, 479…
$ datetime <chr> "30.08.1989 12:00", "30.08.1989 12:00", "30.08.1989 12:00"…
$ latitude <dbl> 66.4, 66.4, 66.4, 66.4, 66.4, 66.4, 66.4, 66.4, 66.4, 66.4…
$ longitude <dbl> -18.8333, -18.8333, -18.8333, -18.8333, -18.8333, -18.8333…
$ pressure <dbl> 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,…
$ temperature <dbl> 7.16, 7.16, 7.16, 7.16, 7.16, 7.13, 7.13, 7.13, 7.14, 7.14…
$ salinity <dbl> 33.160, 33.173, 33.153, 33.152, 33.142, 33.146, 33.144, 33…
$ name <chr> "SI2", "SI2", "SI2", "SI2", "SI2", "SI2", "SI2", "SI2", "S…q |>
collect() |>
ggplot(aes(temperature, -pressure)) +
theme_bw() +
geom_point(size = 0.1, alpha = 0.05) +
labs(title = "Temperature profiles")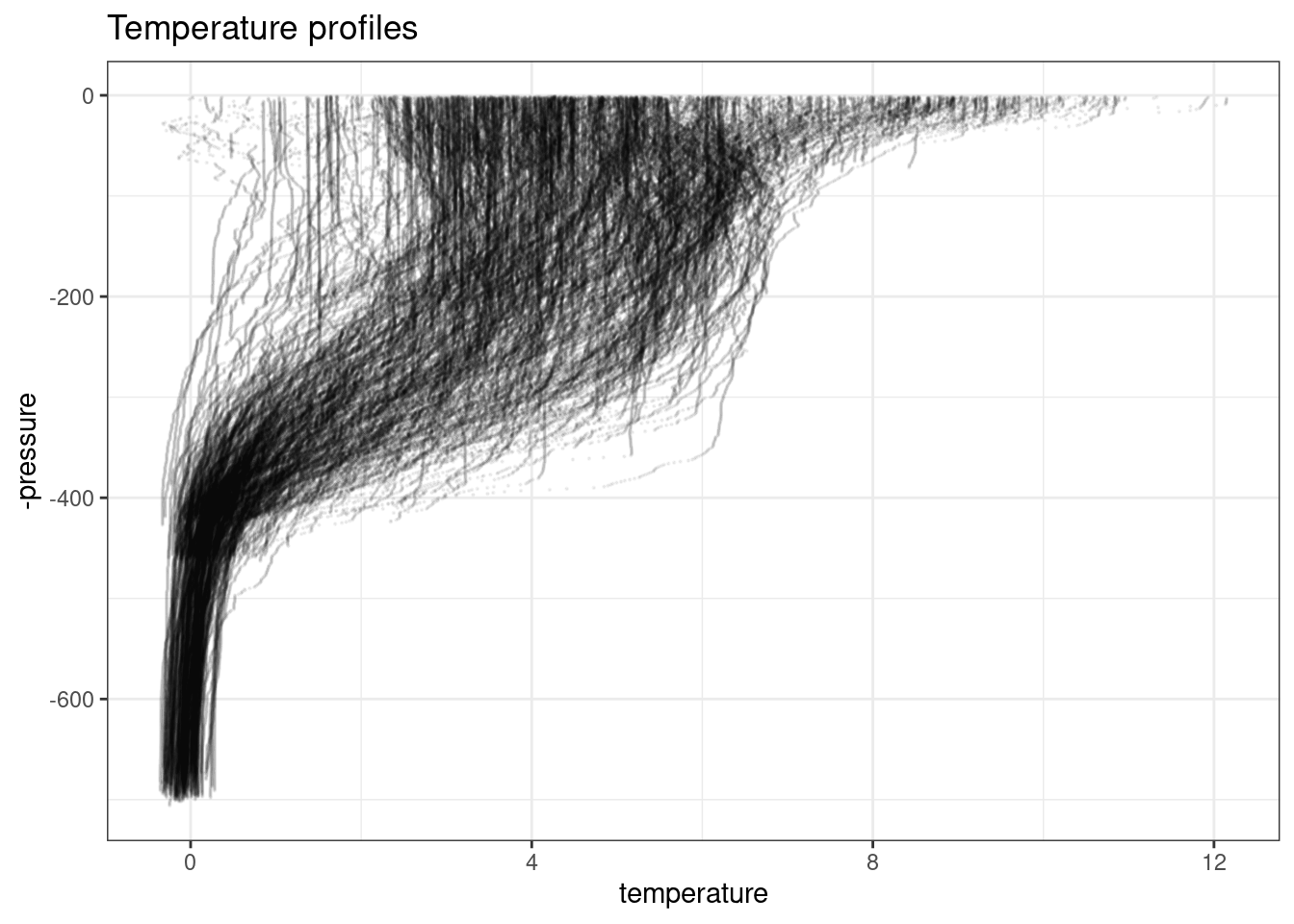
q |>
filter(between(pressure, 150, 200)) |>
mutate(year = str_sub(datetime, 7, 10),
year = as.integer(year)) |>
ggplot(aes(year, temperature)) +
stat_summary(fun.data = "mean_cl_boot") +
labs(title = "Temperature change at pressure 150-200")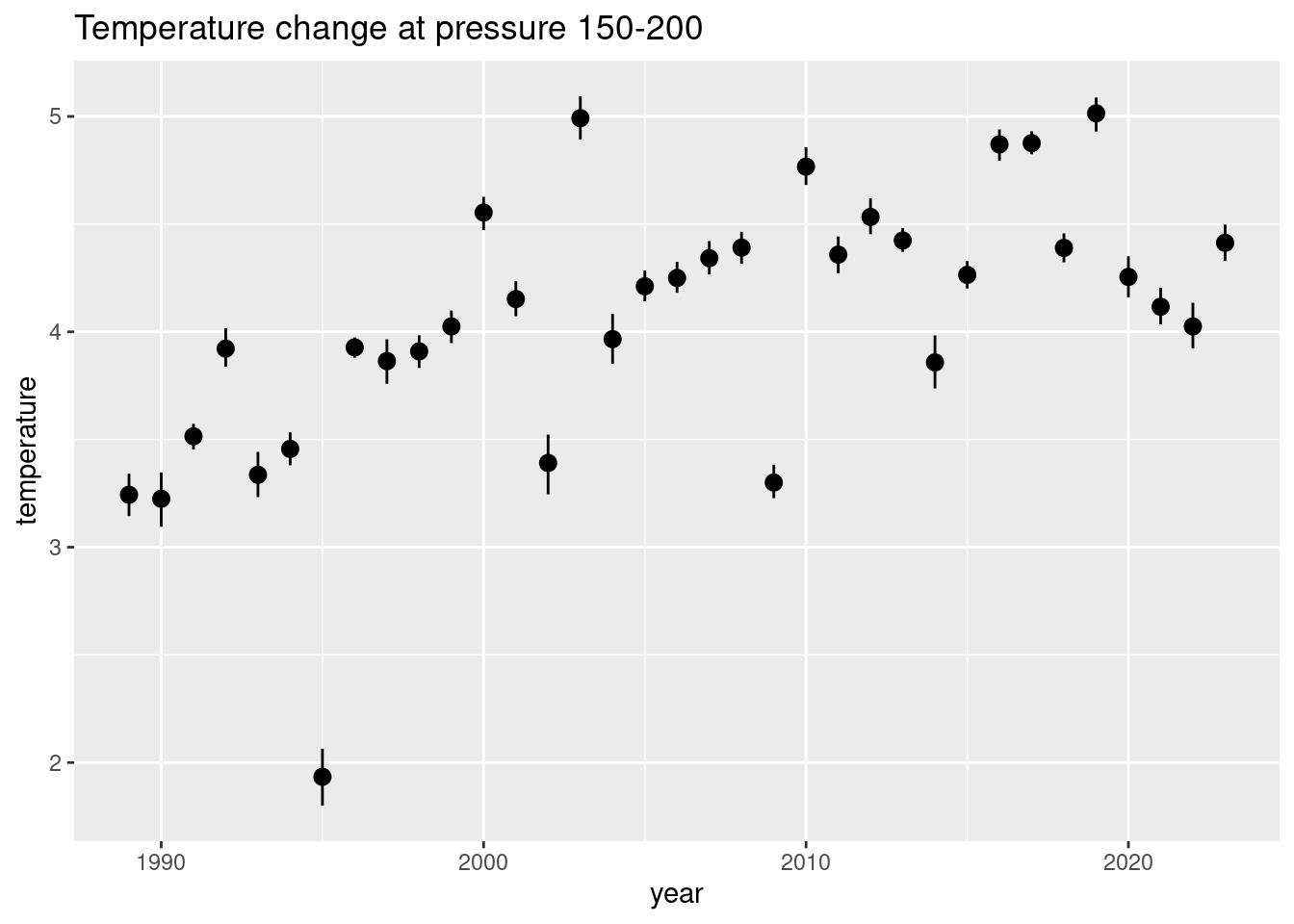
tbl_mar(con, "veidibok.veidibok_v") |>
filter(vatnsfall %in%
c("Laxá í Aðaldal", "Laxá á Ásum,", "Vatnsdalsá", "Ytri Rangá",
"Víðidalsá og Fitjá", "Eystri Rangá", "Norðurá Borg", "Apavatn",
"Langá", "Hlíðarvatn", "Arnarvatn stóra", "Grímsá Borgarfirði",
"Miðfjarðará", "Blanda", "Laxá í Kjós", "Hofsá í Vopnafirði",
"Selá í Vopnafirði", "Fljótaá", "Eyjafjarðará", "Grenlækur")) |>
mutate(year = year(dags)) |>
count(year, vatnsfall, fisktegund) |>
collect() |>
ggplot(aes(year, n, fill = fisktegund)) +
geom_col() +
facet_wrap(~ vatnsfall, scales = "free_y", ncol = 2) +
scale_fill_brewer(palette = "Set1") +
labs(title = "Fjöldi fiska??")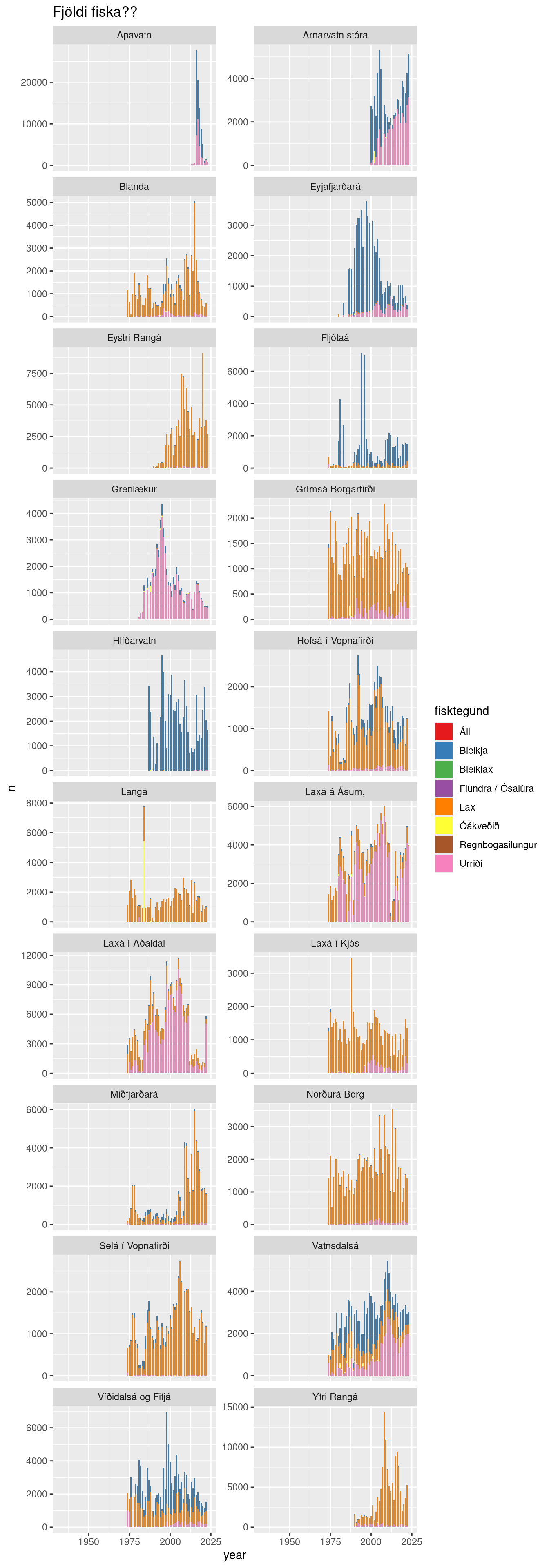
The following shows you can generate very complex query from within R. Here we connect to different tables and then join them using only R-syntax. This is actually the code behind the taggart-funtion used above.
stodvar <-
les_stod(con) %>%
left_join(les_syni(con), by = 'stod_id') %>%
# dplyr::mutate(tLon = kastad_lengd,
# tLat = kastad_breidd,
# tAr = to_number(to_char(dags, 'yyyy'))) %>%
dplyr::select(synis_id,
leidangur,
stod = stod_id,
tDags = dags,
tAr = ar,
tReitur = reitur,
tSmareitur = smareitur,
tLon = kastad_lengd,
tLat = kastad_breidd,
tDypi = botndypi_kastad,
tVeidarfaeri = veidarfaeri)
fiskar <-
fiskmerki_fiskar(con) %>%
dplyr::select(fiskur_id,
synis_id,
tTegund = tegund,
tLengd = lengd,
tThyngd = thyngd,
tKyn = kyn,
tKynthroski = kynthroski)
merki <-
fiskmerki_merki(con) %>%
dplyr::select(tid,
fiskur_id,
audkenni,
numer)
q <-
fiskar %>%
dplyr::left_join(merki, by = "fiskur_id") %>%
dplyr::left_join(stodvar, by = "synis_id") %>%
dplyr::left_join(fiskmerki_endurheimtur(con), by = "tid") %>%
dplyr::left_join(fiskmerki_rafaudkenni(con), by = "tid") %>%
dplyr::select(-id, -tid)
q |> show_query()