intro
intro.Rmd
library(sf)
library(boot)
library(broom)
library(dplyr)
library(tidyr)
library(purrr)
library(broom)
library(ggplot2)
library(pgnapes)
con <- pgn_connect(username = "your_user_name", password = "your_password")
pgnapes::WGINORPolygons %>%
ggplot() +
theme_bw() +
geom_sf(aes(colour = region),
alpha = 0, lwd = 1)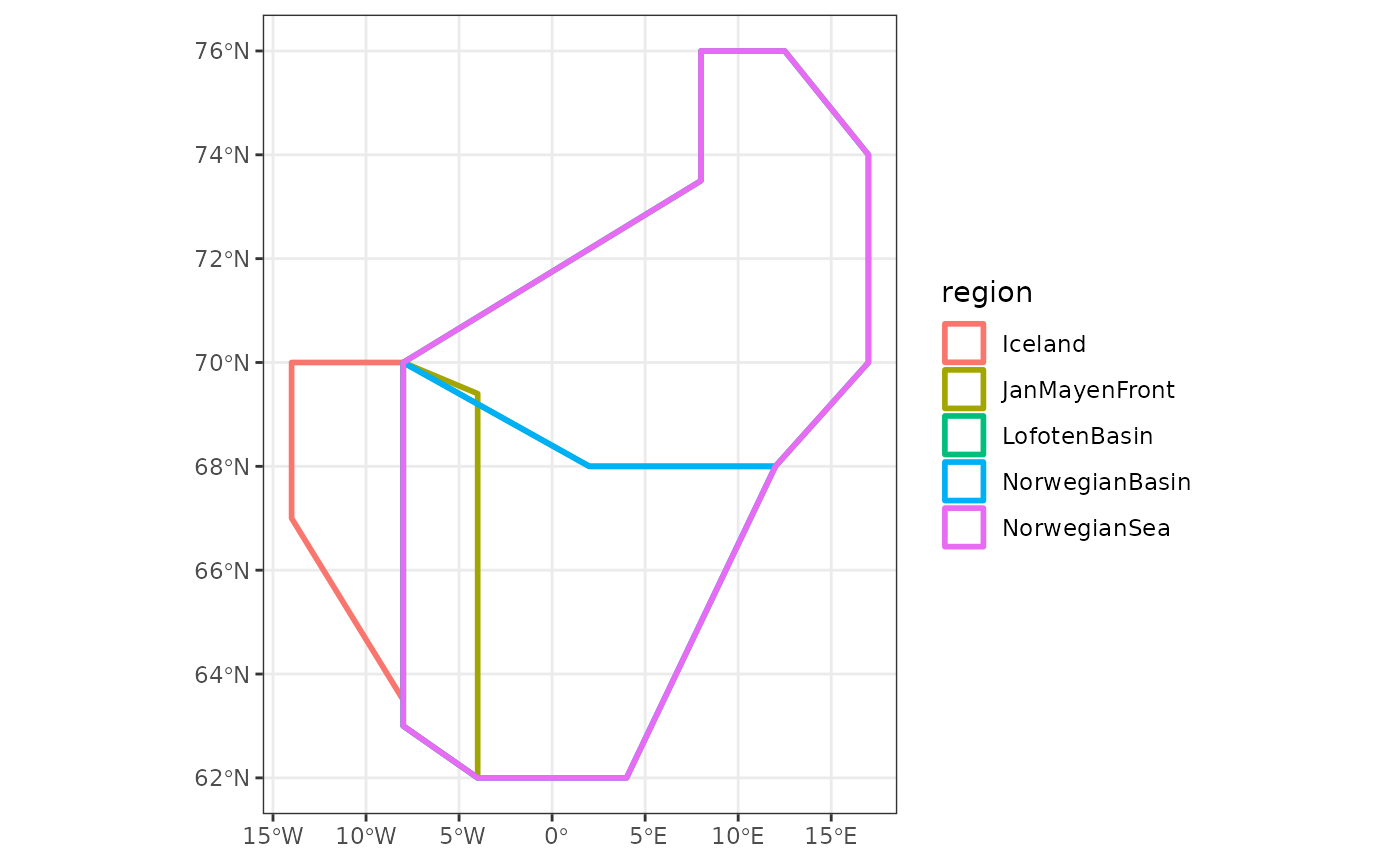
hmmm … the overlap of polygons may create problem downstream
pgn_plankton(con) %>%
select(country:year, sumdrywt) %>%
left_join(pgn_logbook(con) %>%
select(country:year, month, lon, lat)) %>%
collect(n = Inf) %>%
st_as_sf(coords = c("lon", "lat"),
crs = 4326) %>%
st_join(WGINORPolygons %>%
filter(region != "NorwegianSea")) %>%
st_drop_geometry() %>%
count()
#> # A tibble: 1 × 1
#> n
#> <int>
#> 1 8126
# problem is the shapefile, hence more rows than in the original data
d <-
pgn_plankton(con) %>%
select(country:year, sumdrywt) %>%
left_join(pgn_logbook(con) %>%
select(country:year, month, lon, lat)) %>%
collect(n = Inf) %>%
st_as_sf(coords = c("lon", "lat"),
crs = 4326) %>%
st_join(WGINORPolygons %>%
filter(region %in% c("Iceland", "JanMayenFront",
"LofotenBasin", "NorwegianBasin"))) %>%
st_drop_geometry() %>%
filter(!is.na(region))
d2 <-
d %>%
filter(year >= 2010) %>%
select(year, region, sumdrywt) %>%
group_by(year, region) %>%
nest() %>%
mutate(boot_res = map(data,
~ boot(data = .$sumdrywt,
statistic = function(x, i) mean(x[i]),
R = 1000)),
boot_tidy = map(boot_res, tidy, conf.int = TRUE, conf.method = "perc"),
n = map(data, nrow)) %>%
select(-data, -boot_res) %>%
unnest(cols = -c(year, region)) %>%
ungroup()
d2 %>% arrange(region, year)
#> # A tibble: 52 × 8
#> year region statistic bias std.error conf.low conf.high n
#> <dbl> <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <int>
#> 1 2010 Iceland 4860. 12.7 463. 3994. 5884. 48
#> 2 2011 Iceland 5907. 17.9 569. 4823. 7117. 51
#> 3 2012 Iceland 6489. -22.5 573. 5352. 7600. 70
#> 4 2013 Iceland 9877. 30.2 959. 8044. 11867. 31
#> 5 2014 Iceland 9490. -2.81 891. 7822. 11292. 52
#> 6 2015 Iceland 7719. -2.65 730. 6391. 9160. 50
#> 7 2016 Iceland 7827. -16.5 905. 6114. 9693. 30
#> 8 2017 Iceland 10919. -58.7 1389. 8696. 14210. 24
#> 9 2018 Iceland 9795. -64.6 1604. 6884. 13174. 26
#> 10 2019 Iceland 7582. -3.28 756. 6156. 9094. 39
#> # … with 42 more rows
d2 %>%
ggplot(aes(year, statistic)) +
geom_ribbon(aes(ymin = conf.low,
ymax = conf.high),
fill = "pink") +
geom_point() +
geom_line() +
facet_wrap(~ region, scales = "free_y") +
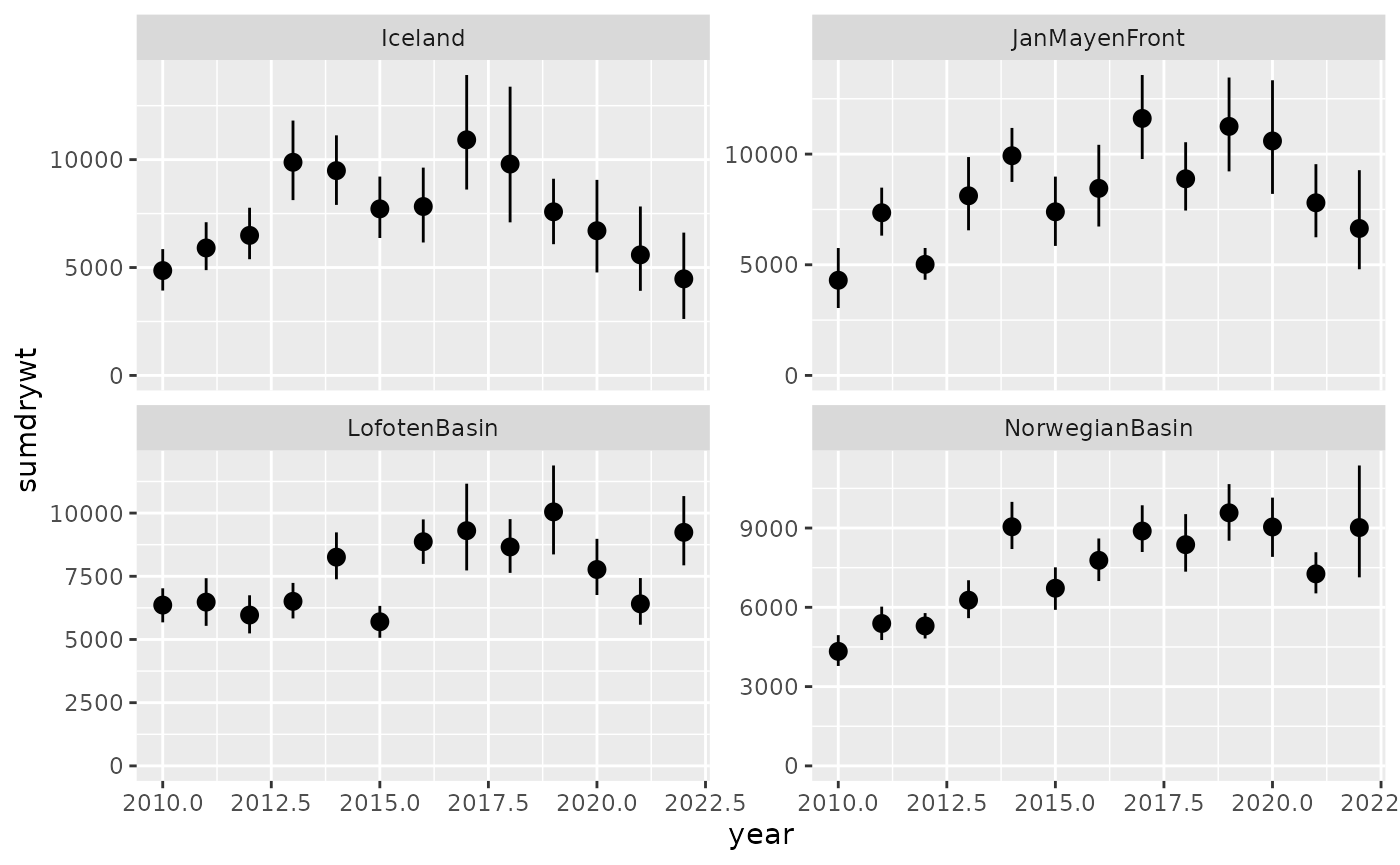
expand_limits(y = 0)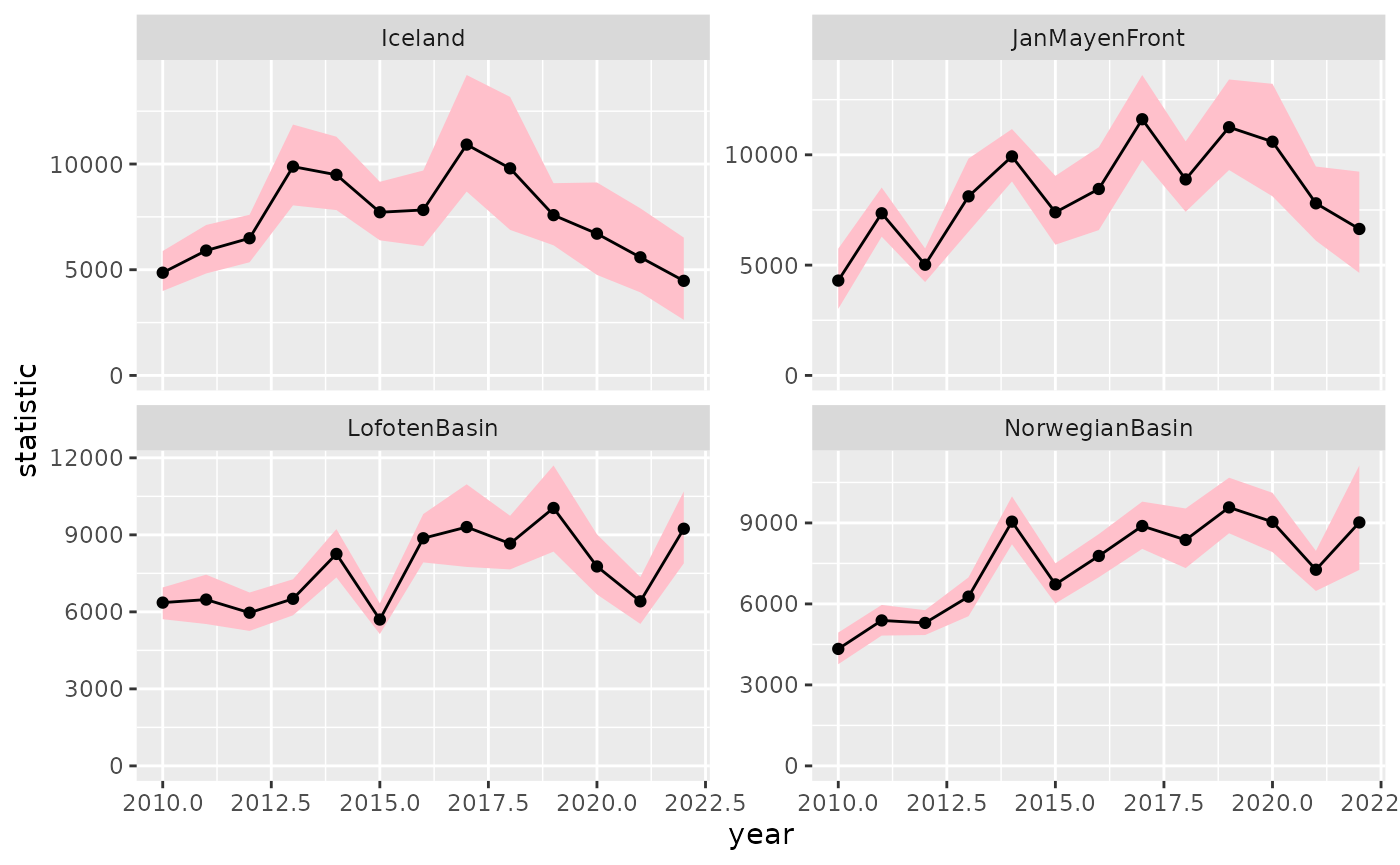
Same could be achieved as plot only via:
d %>%
filter(year >= 2010) %>%
ggplot(aes(year, sumdrywt)) +
stat_summary(fun.data = "mean_cl_boot") +
facet_wrap(~ region, scales = "free_y") +
expand_limits(y = 0)