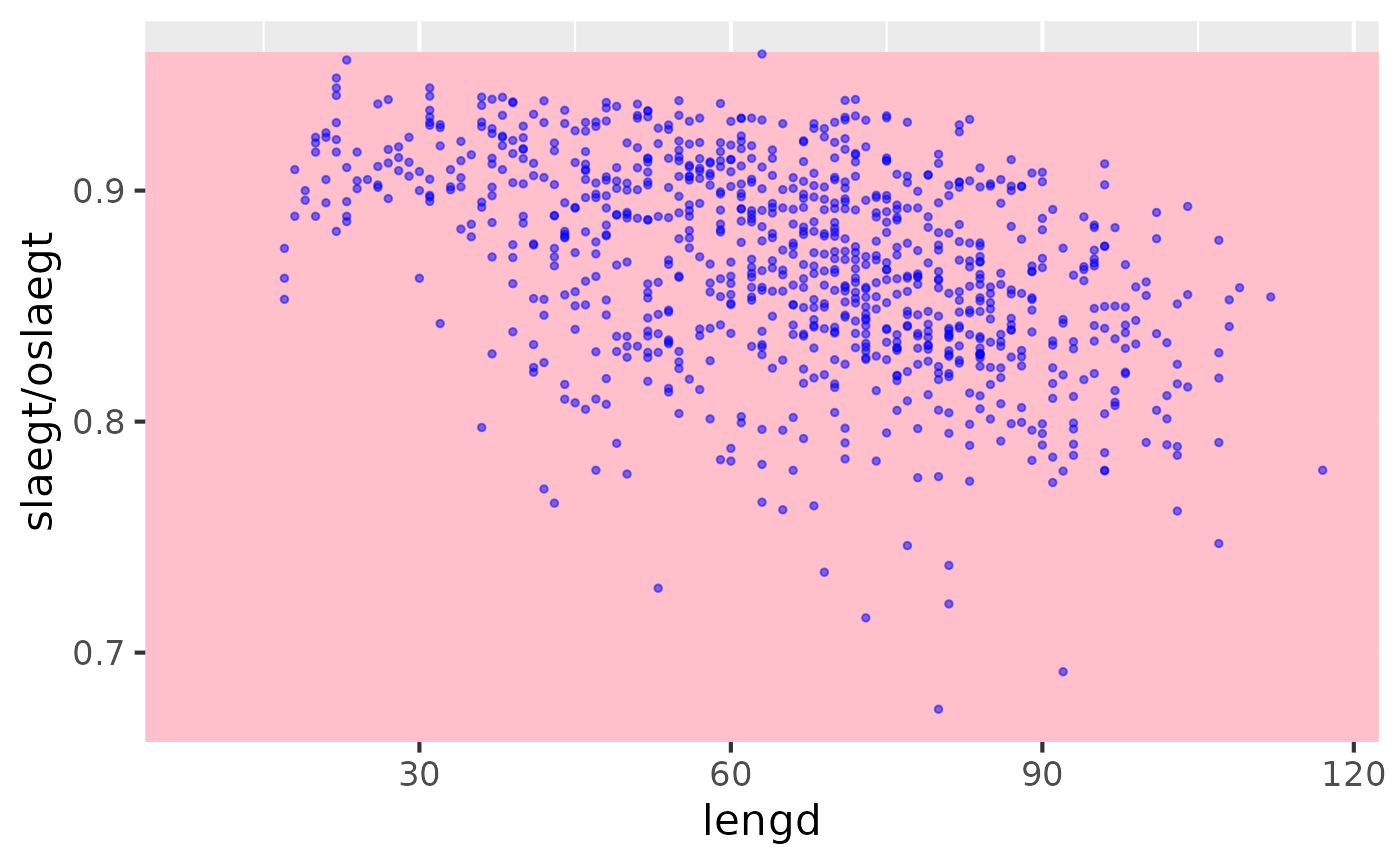
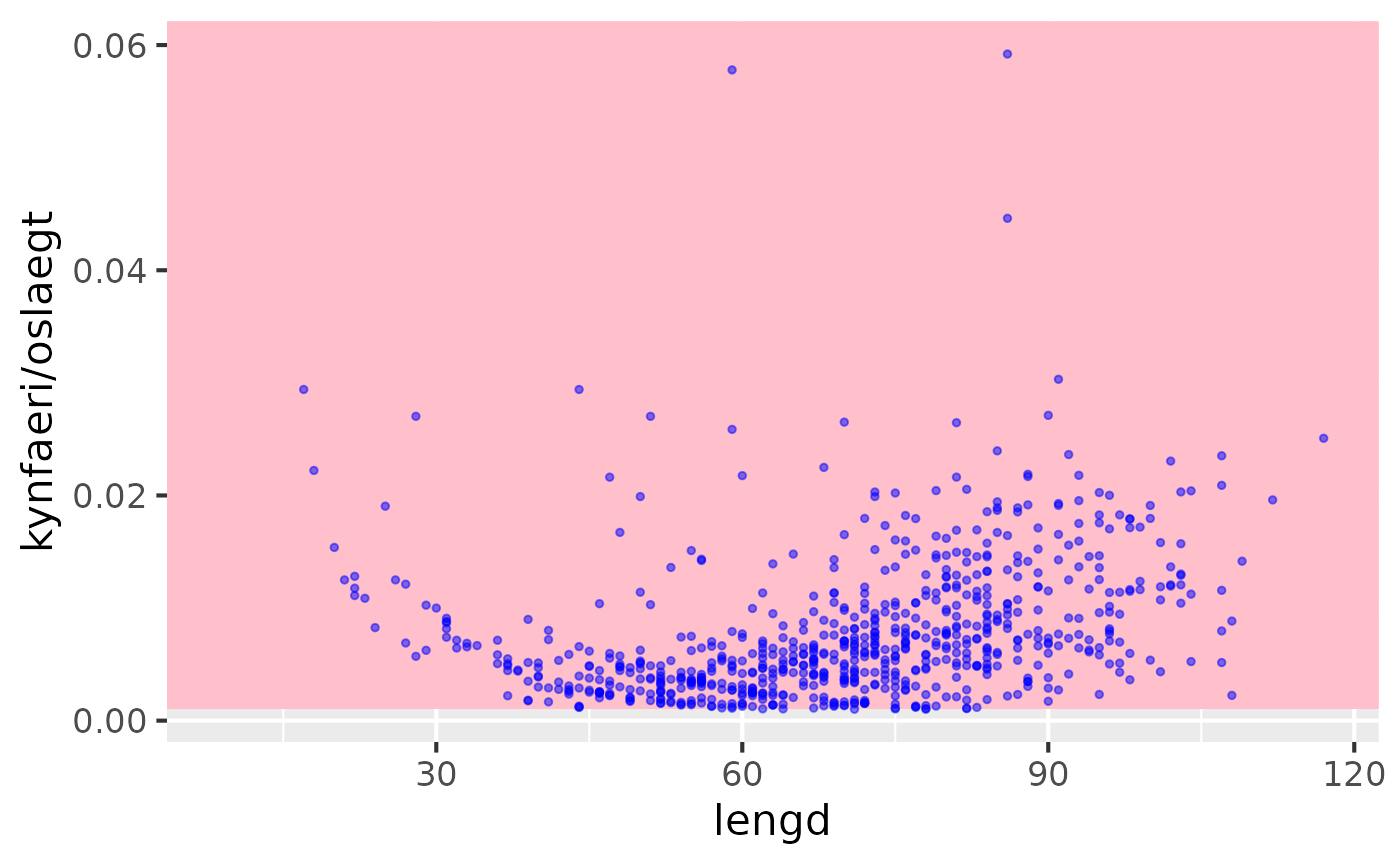
demo
emulation.RmdReading data from dumped zip-files
One can read in the data currently being collected using the
hv_import_cruise that resides in the {ovog}-package:
cruises <- here::here(c("data-raw/TB2-2024.zip", "data-raw/TTH1-2024.zip"))
measurement <-
ovog::hv_import(cruises) |>
ovog::hv_create_tables()The object returned is a list containing the following tables:
names(measurement)
#> [1] "stodvar" "skraning" "numer" "lengdir" "kvarnir" "pp"Explorations - A sidestep
There should be nothing stopping you from doing analysis of the this data, e.g. one can easily get an overview of the reported station location via:
measurement$stodvar |>
ggplot() +
theme_bw() +
geom_segment(aes(x = -kastad_v_lengd, y = kastad_n_breidd,
xend = -hift_v_lengd, yend = hift_n_breidd,
colour = factor(fishing_gear_no)),
linewidth = 1) +
coord_quickmap()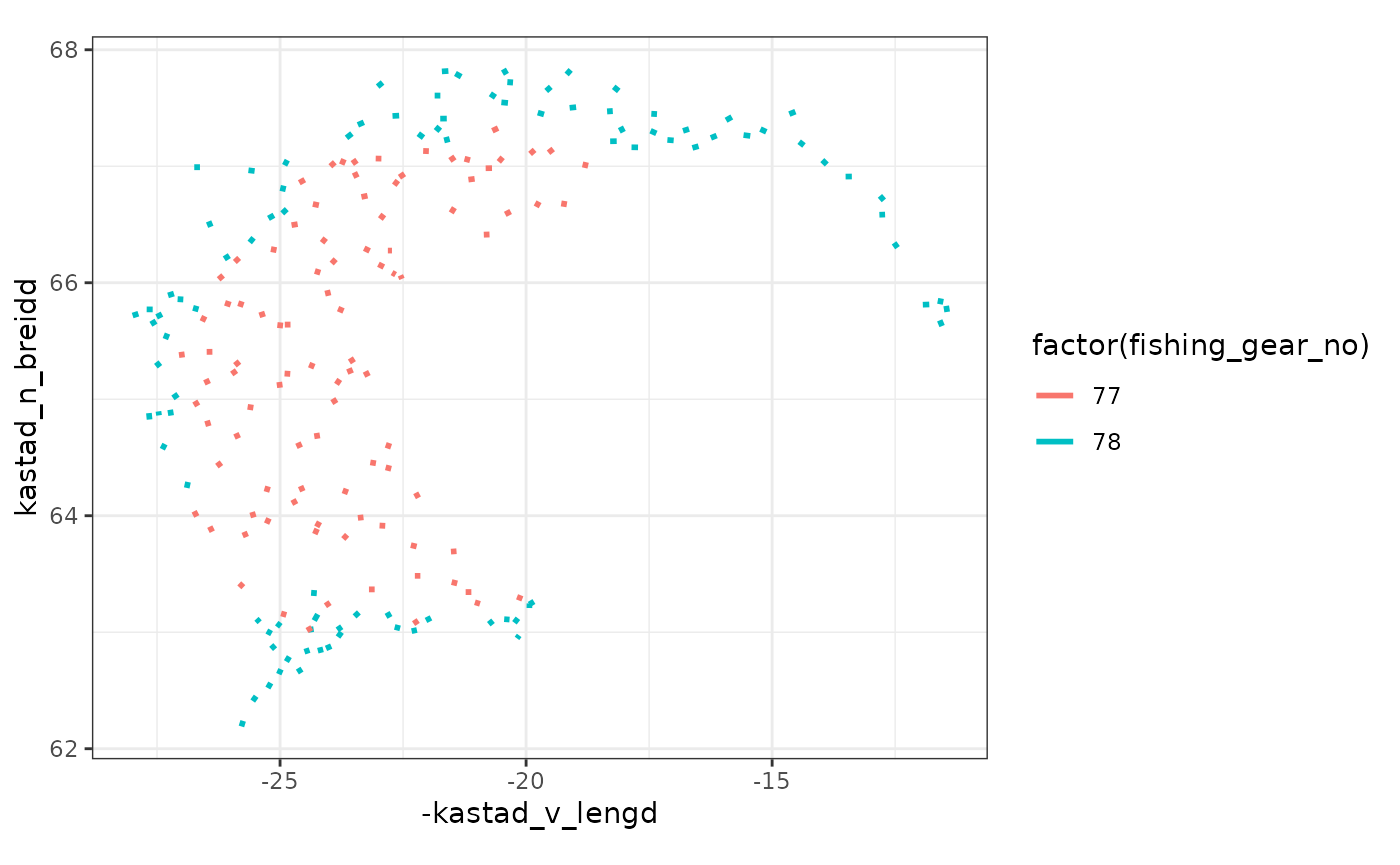
Or just the length distributions of some selected species via:
measurement$lengdir |>
filter(tegund %in% 1:3) |>
group_by(tegund, lengd) |>
reframe(n = sum(n)) |>
ggplot(aes(lengd, n)) +
geom_col() +
facet_wrap(~ tegund, scales = "free_y")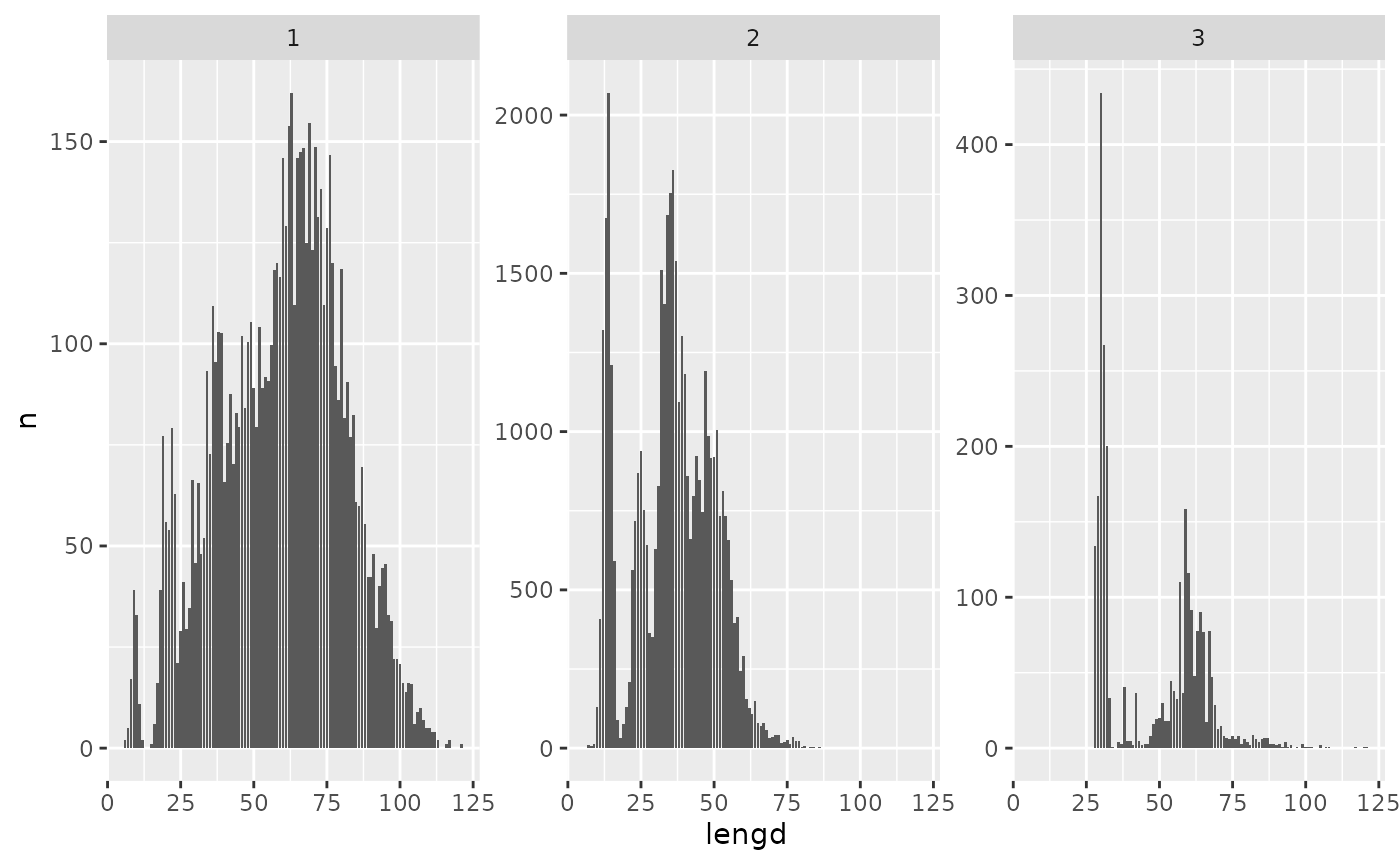
Merging and data transformation
Normally we would …
cruises <- here::here(c("data-raw/TB2-2024.zip", "data-raw/TTH1-2024.zip"))
stillingar <- here::here("data-raw/stillingar_SMH_rall_(haust).zip")
stodtoflur <- here::here("data-raw/stodtoflur.zip")
res <- sm_munge(cruises, stillingar, stodtoflur)
#> IMPORT
#> Import current measurements
#> The 'synaflokkur' of the zip files is: 35
#> Import setup ('stillingar')
#> Rename variables in sti_rallstodvar
#> Import setup ('stodtoflur')
#> Importing historical measurements (takes a while)
#> Combine current and historical data
#> MUNGE
#> Scale by counted
#> Standardize to towlength
#> Results by year and length
#> Results by station
#>
#> Trawl metrics and temperature
#> Time series
#> Last 20 tows
#> Tidy 'Stillingar'
#> 'Length-weight'
#> Checking measurments
#> Checking length vs weights
#> Checking gutted, gonads, liver vs ungutted ratio
#> REMINDER: Need to get data on magi
#> Formatting QC table
#> Togfar
#> Handbokartog
#> Valblod
#> Bootstrapping abundance
#>
#> Bootstrapping biomass
#>
#> HURRA! Exploration outside the smx-app
Sum of catch by lengths
res$by.length |>
dplyr::filter(tegund == 1,
ar >= 2010) |>
dplyr::filter(var == "n") |>
sm_plot_lengths()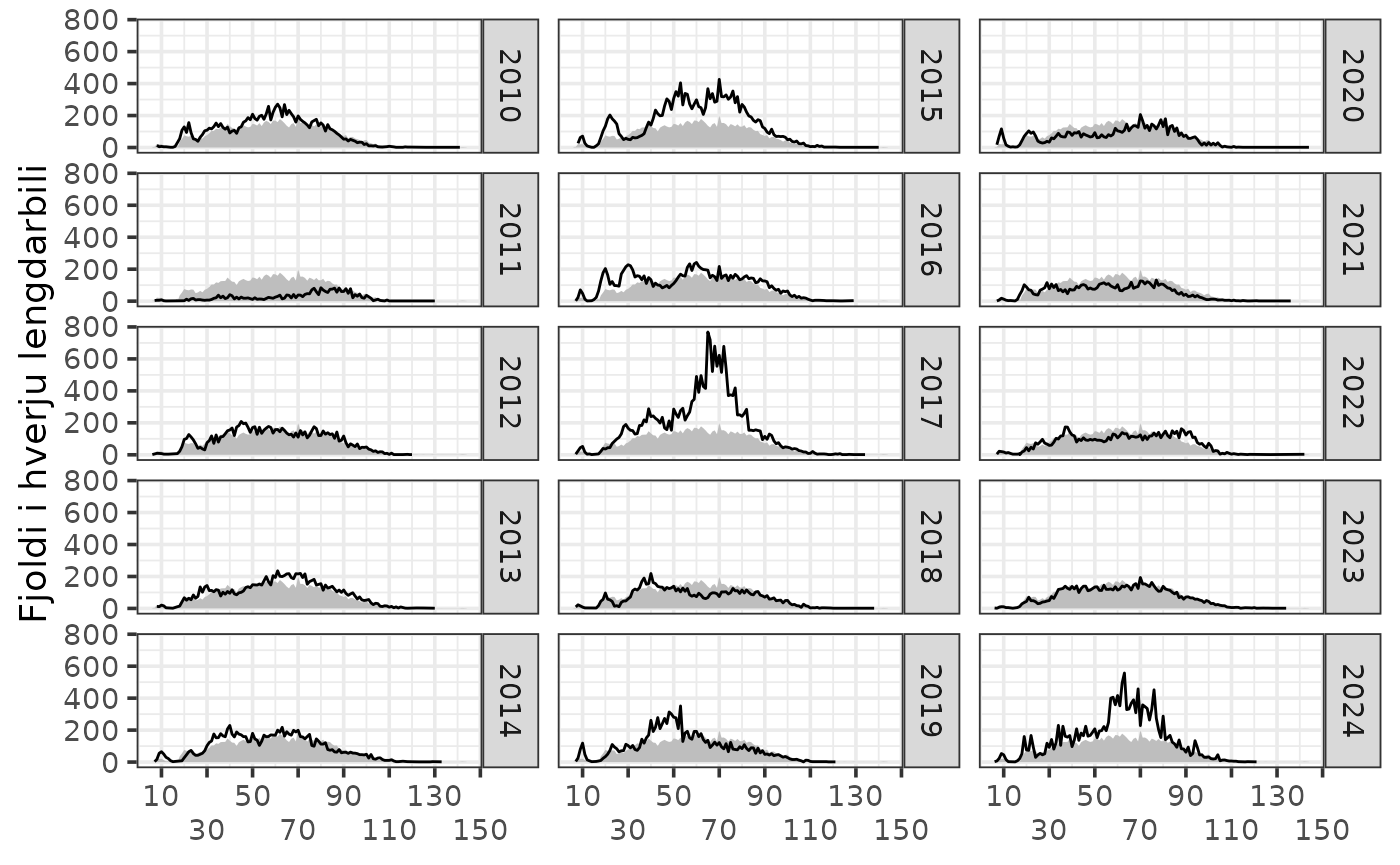
Catch by station
res$by.station |>
dplyr::filter(tegund == 1,
var == "b",
ar %in% c(seq(2000, 2015, by = 5),
2017:2024)) |>
sm_plot_bubble()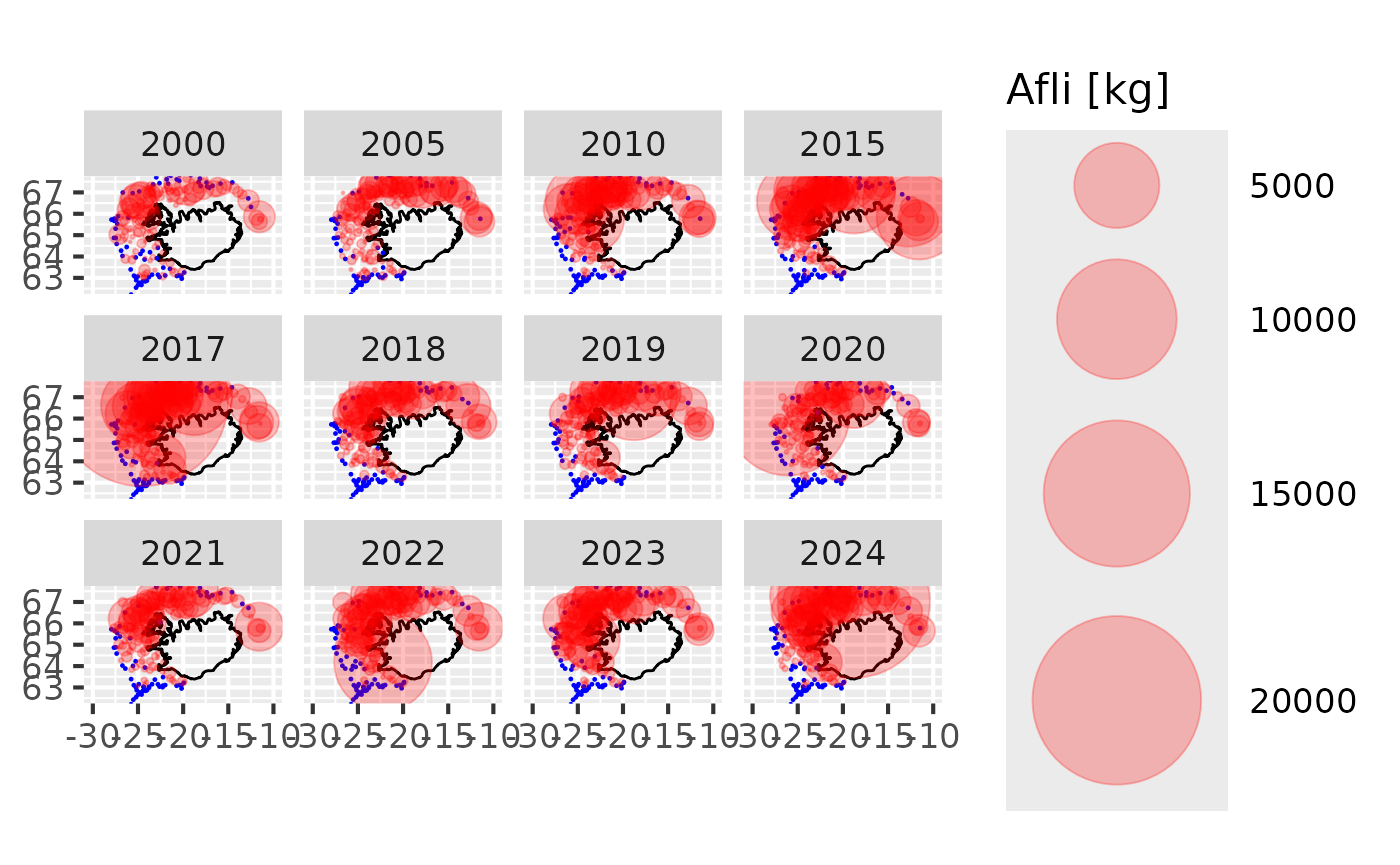
Catch by square
res$by.rect |>
dplyr::filter(tegund == 1,
var == "n") |>
sm_plot_glyph()
#> Registered S3 method overwritten by 'GGally':
#> method from
#> +.gg ggplot2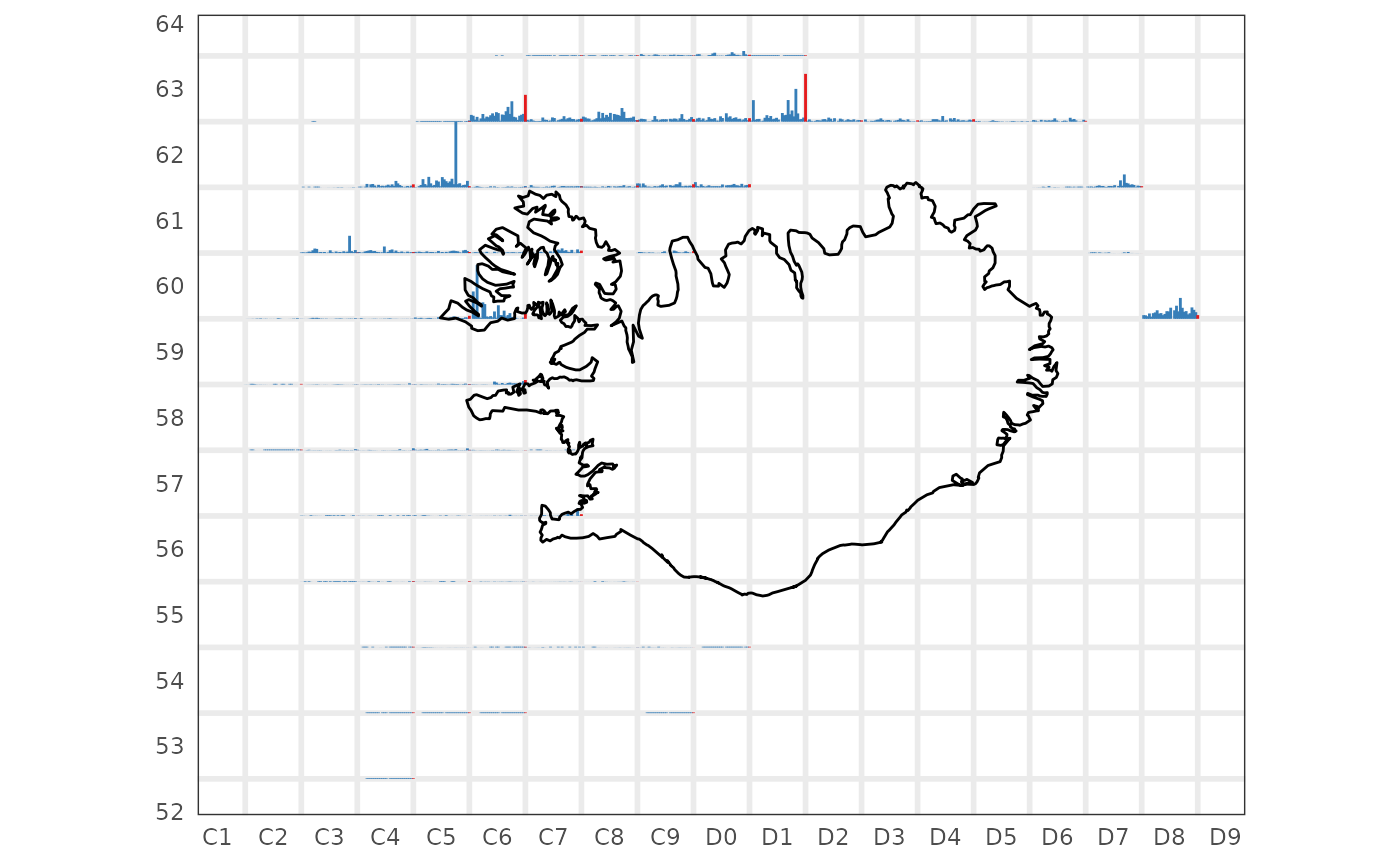
res$by.rect |>
dplyr::filter(tegund == 2,
var == "b") |>
sm_plot_glyph()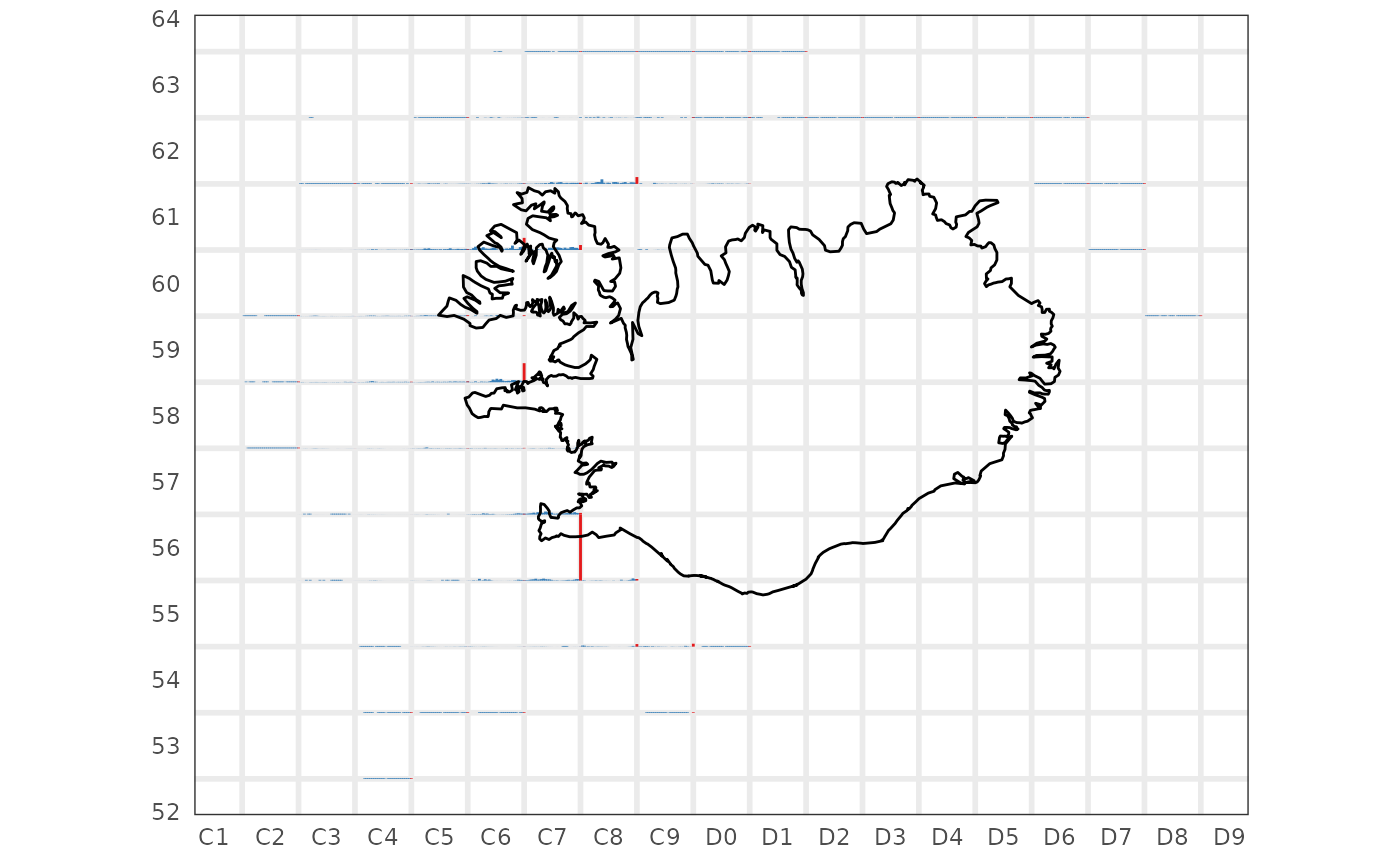
Probability of capture
tmp <-
res$by.station |>
dplyr::filter(ar == max(ar), tegund == 22, var == "n") |>
dplyr::mutate(val = ifelse(val == 0, "absent", "present")) |>
dplyr::select(lon, lat, val)
res$p_capture |>
dplyr::filter(tegund == 22) |>
sm_plot_probability() +
ggplot2::geom_point(data = tmp,
ggplot2::aes(lon, lat, colour = val),
size = 0.5) +
ggplot2::scale_colour_manual(values = c("present" = "cyan", "absent" = "white"),
guide = "none")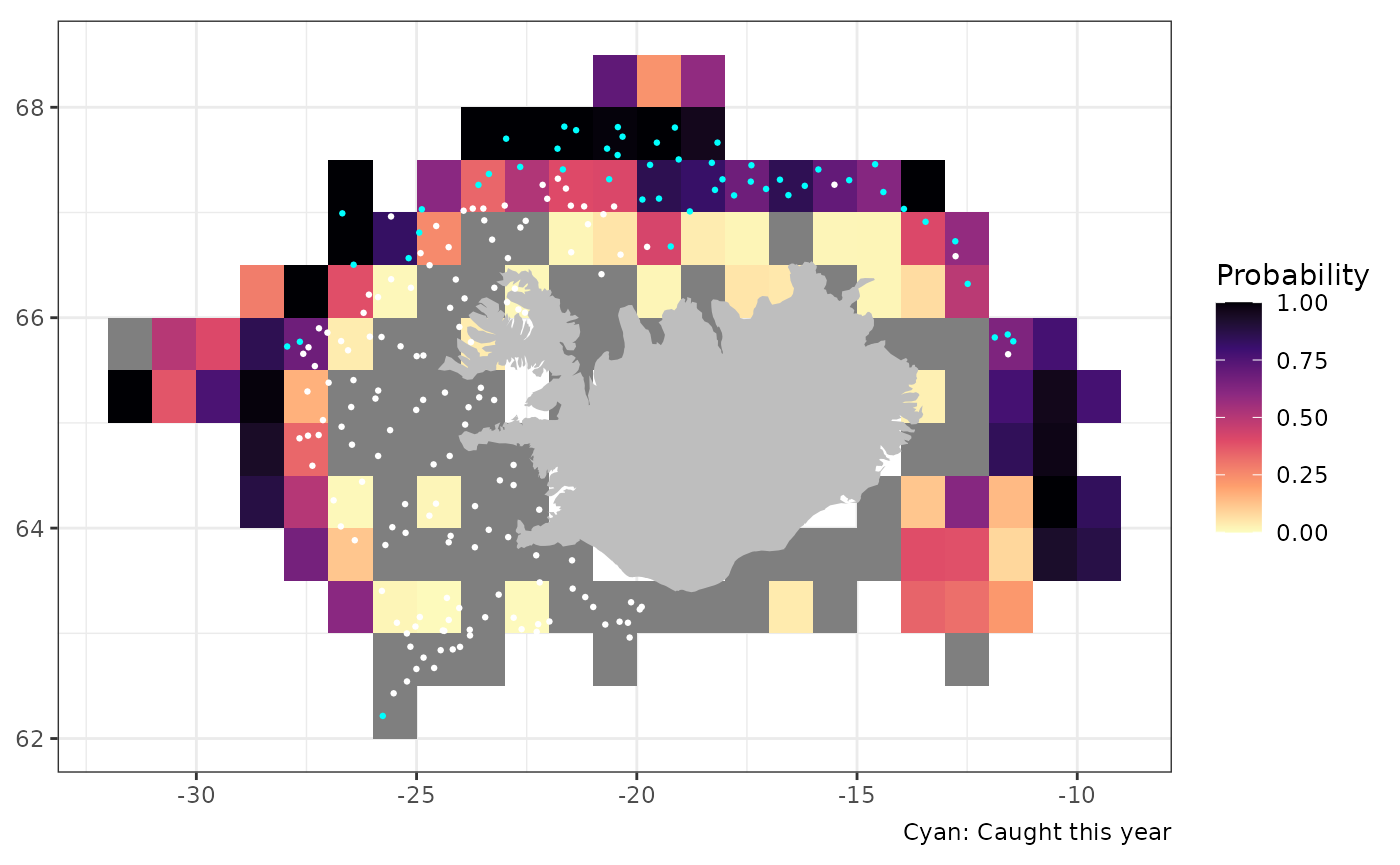
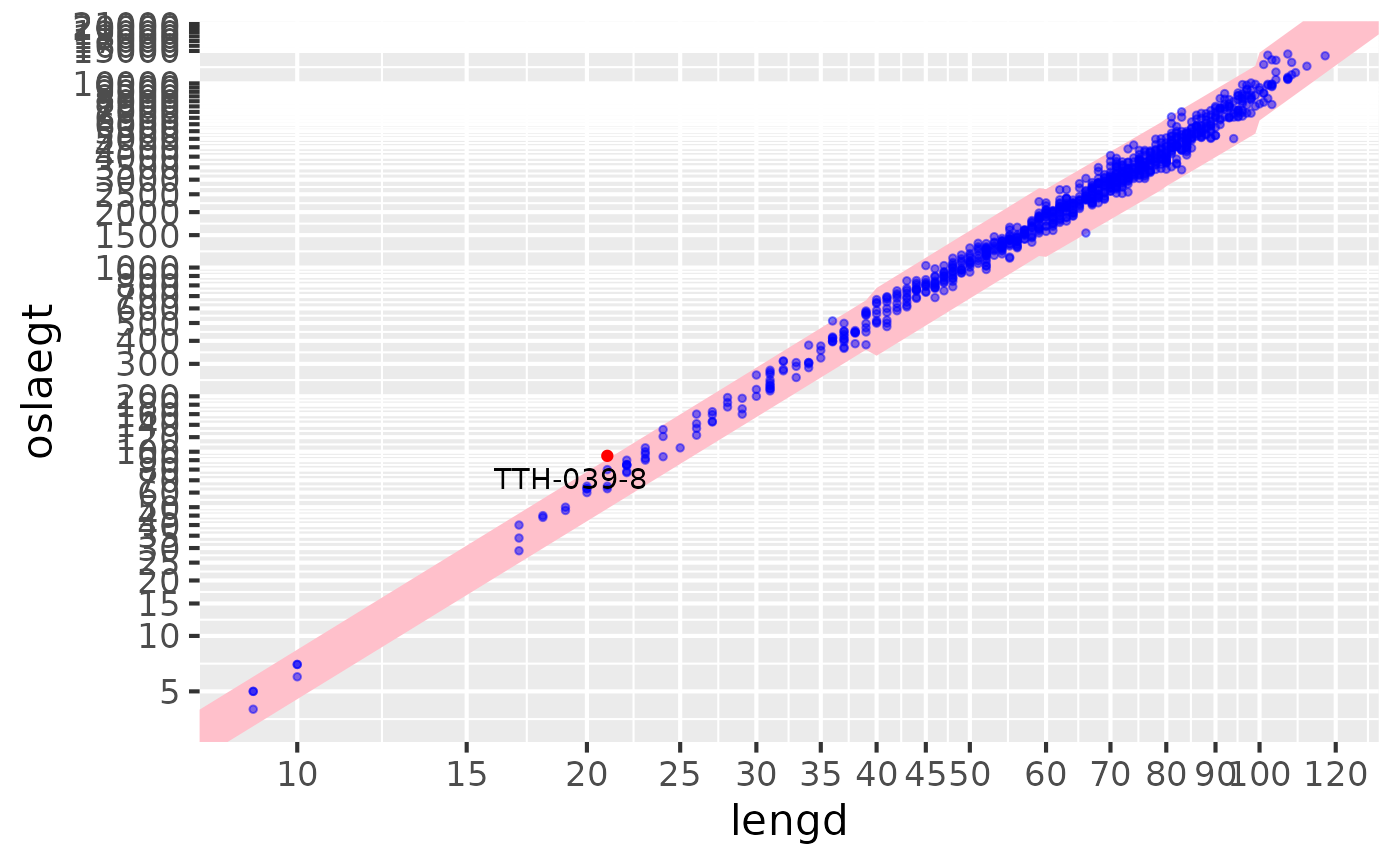
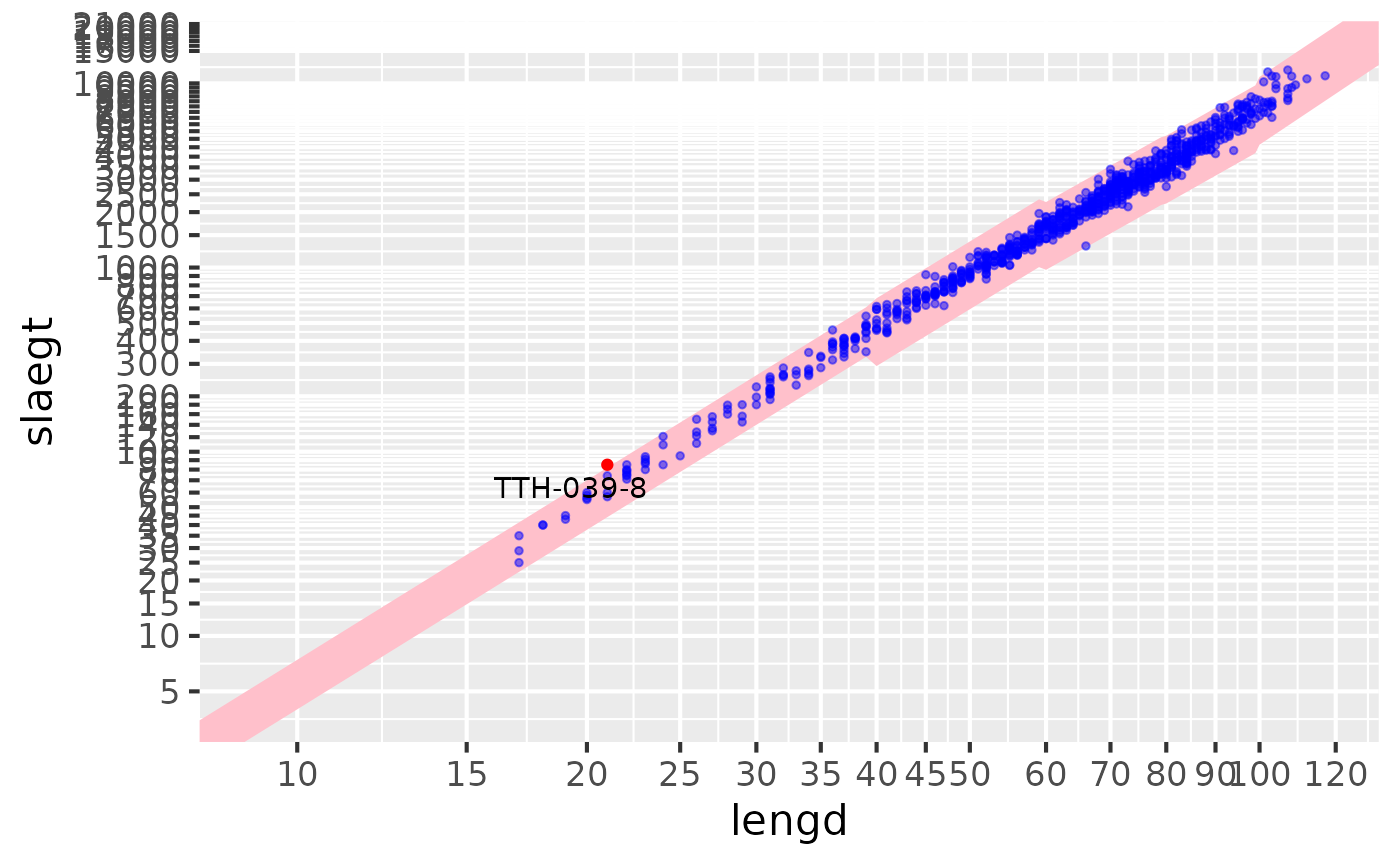
Lengths vs liver / ungutted
res |> sm_plot_liver()
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_point()`).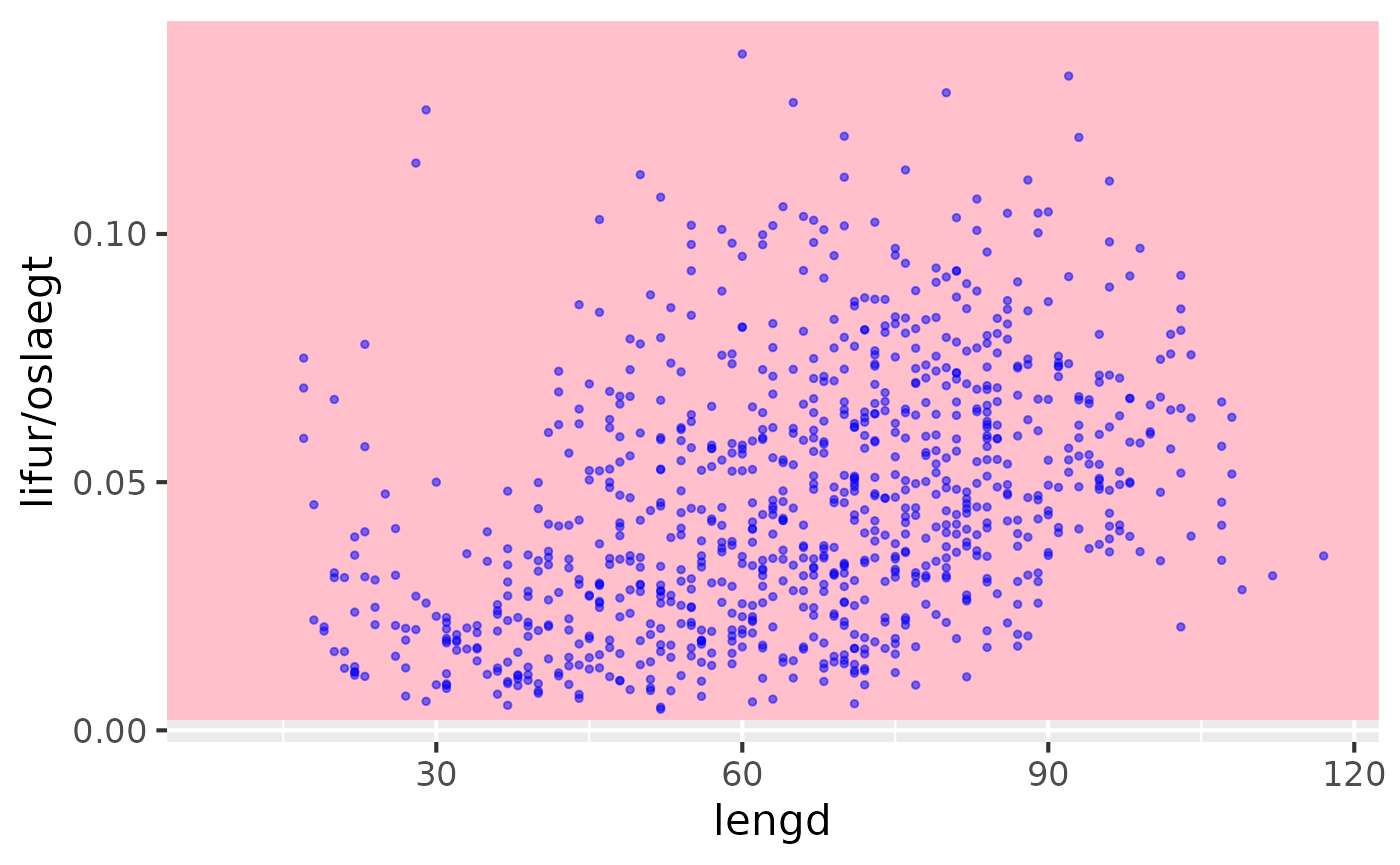
Prey table
res$pp |>
dplyr::filter(astand %in% 1) |>
osmx::sm_table_prey()Prey histogram
### Þyngdardreifing á bráð - Topp 20 bráðir
# applly some filter
pp <-
res$pp |>
dplyr::filter(astand == 1,
# brað verdur ad vera skilgreind
!is.na(prey),
# verður að hafa mælingu thyngd
!is.na(heildarthyngd),
# verður að hafa mælingu talid
!is.na(n)) |>
dplyr::mutate(m.thyngd = round(heildarthyngd / n, 2))
# not really mean based on individual prey measurements
pp.mean <-
pp |>
dplyr::group_by(prey) |>
dplyr::summarise(fjoldi = sum(n),
mean = sum(heildarthyngd) / sum(n)) |>
dplyr::ungroup() |>
dplyr::arrange(-fjoldi)
# top 20 (bráð) tegundir "mældar"
top20 <-
pp.mean |>
dplyr::slice(1:20)
top20.brad <-
top20 |>
dplyr::pull(prey)
pp |>
dplyr::filter(prey %in% top20.brad) |>
ggplot2::ggplot() +
ggplot2::geom_histogram(ggplot2::aes(m.thyngd)) +
ggplot2::geom_vline(data = top20,
ggplot2::aes(xintercept = mean), colour = "red") +
ggplot2::facet_wrap(~ prey, scale = "free") +
ggplot2::labs(x = "Meðalþyngd", y = "Fjöldi")
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.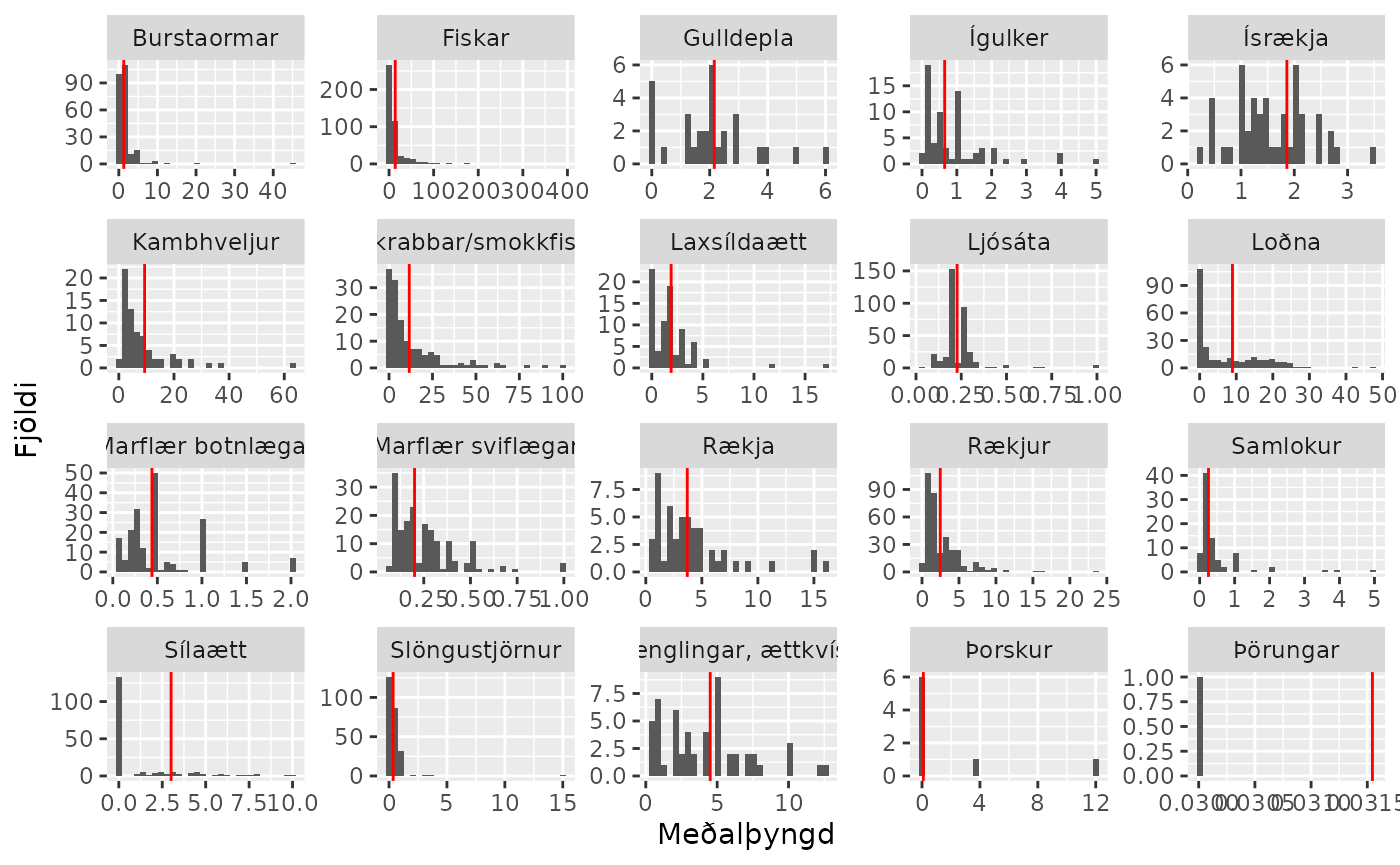
Pred-prey figures
tmp |>
ggplot2::ggplot() +
ggplot2::geom_histogram(ggplot2::aes(magafylli)) +
ggplot2::facet_wrap(~ fiskur, scales = "free")
#> `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.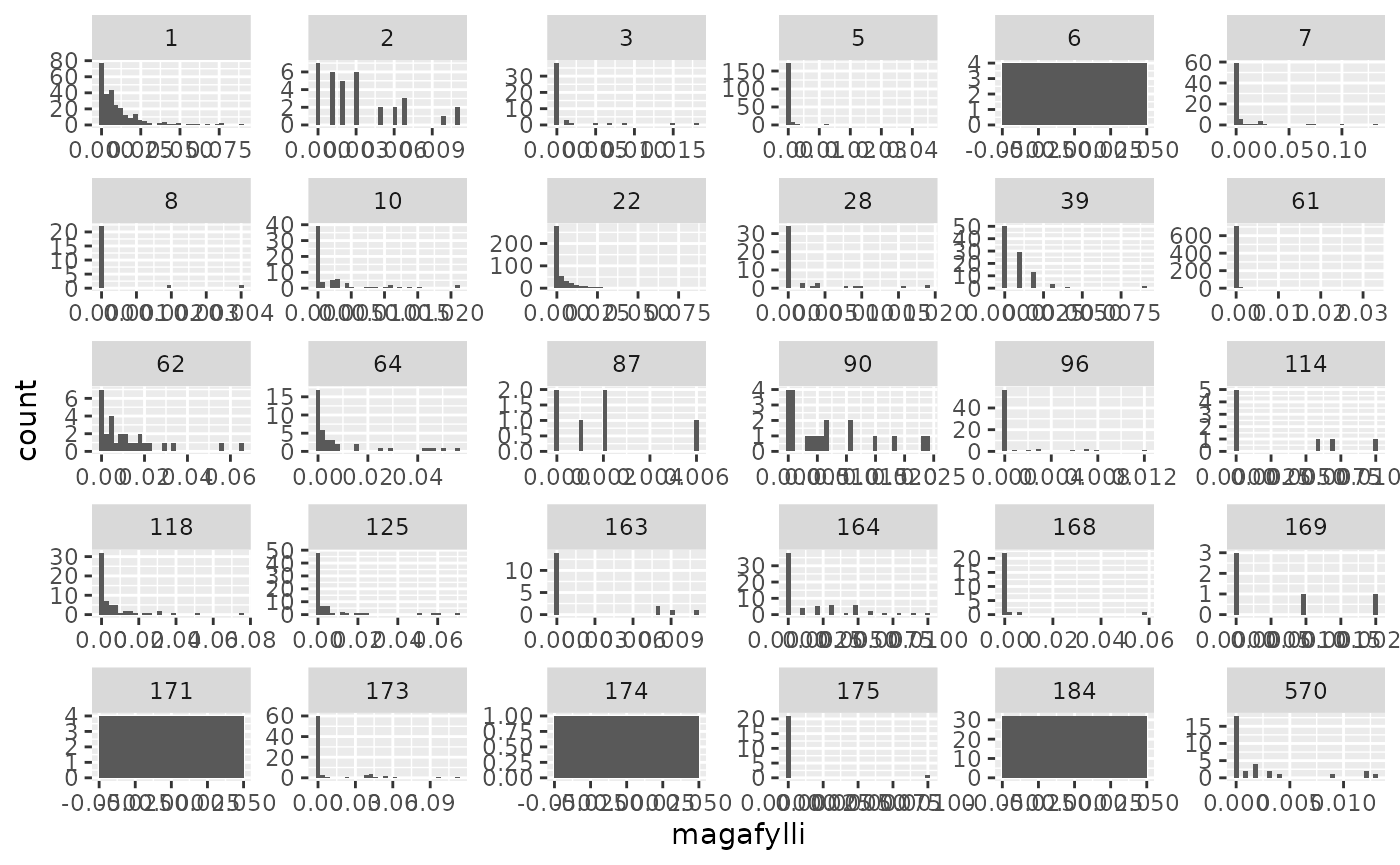
res$kv.this.year |>
dplyr::filter(leidangur %in% "TB2-2024") |>
sm_table_kvarnir()Gear metrics - last 20 stations
res$timetrend.20 |>
filter(leidangur %in% "TB2-2024",
var %in% c("larett_opnun", "lodrett_opnun", "vir_uti")) |>
sm_plot_last20()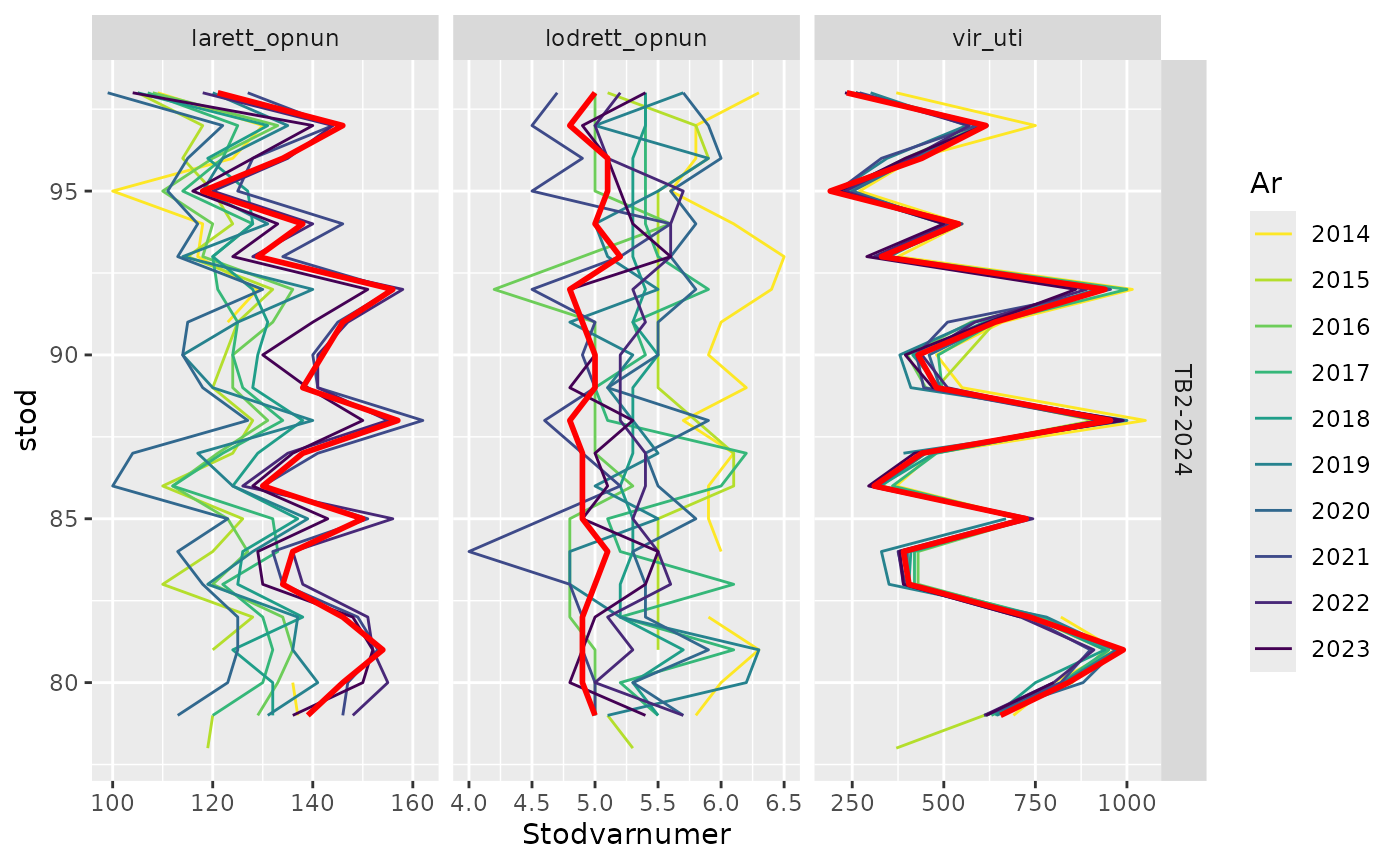
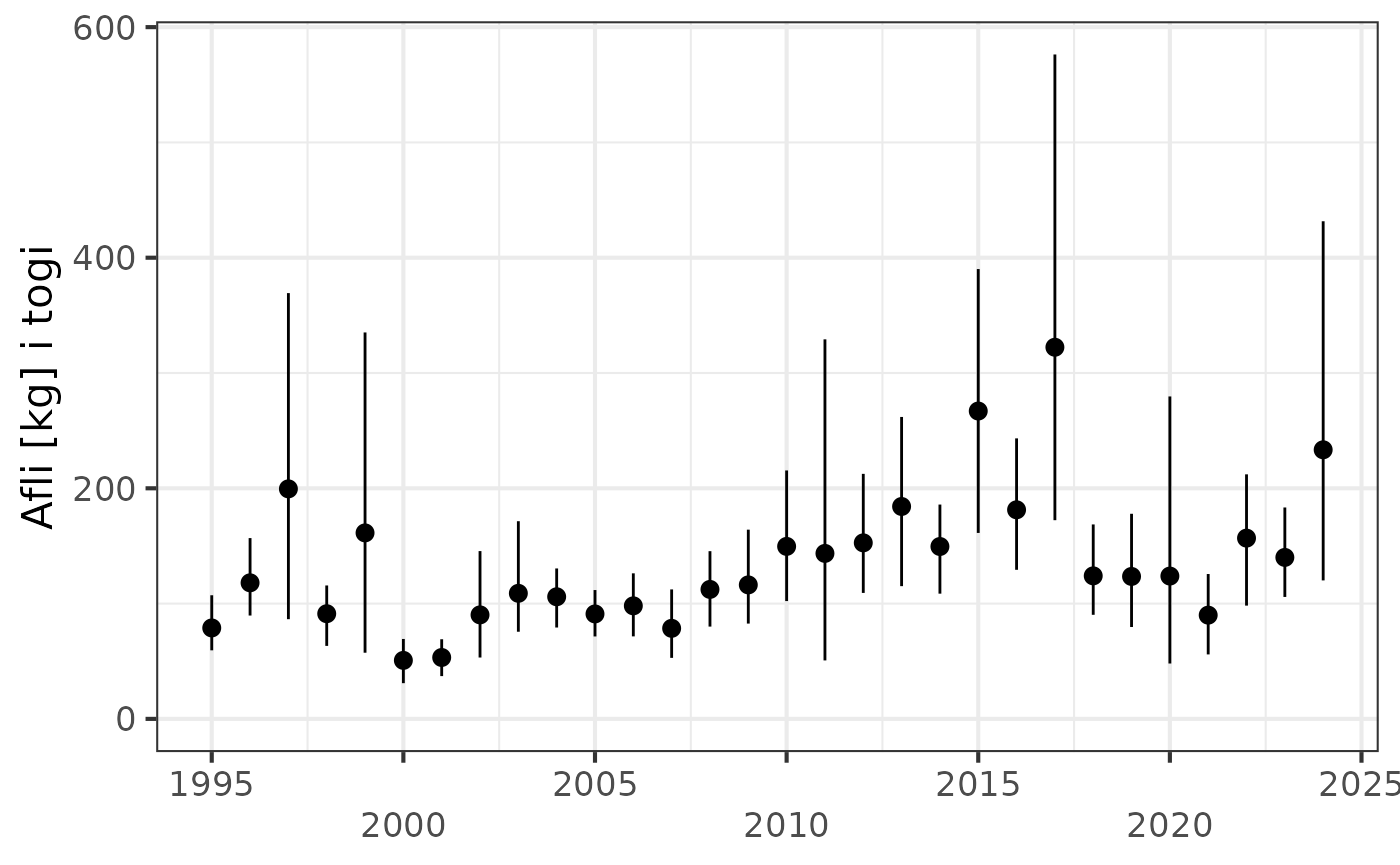
Temperatures - last 20 stations
res$timetrend.20 |>
filter(leidangur %in% "TB2-2024",
var %in% c("botnhiti", "yfirbordshiti")) |>
sm_plot_last20()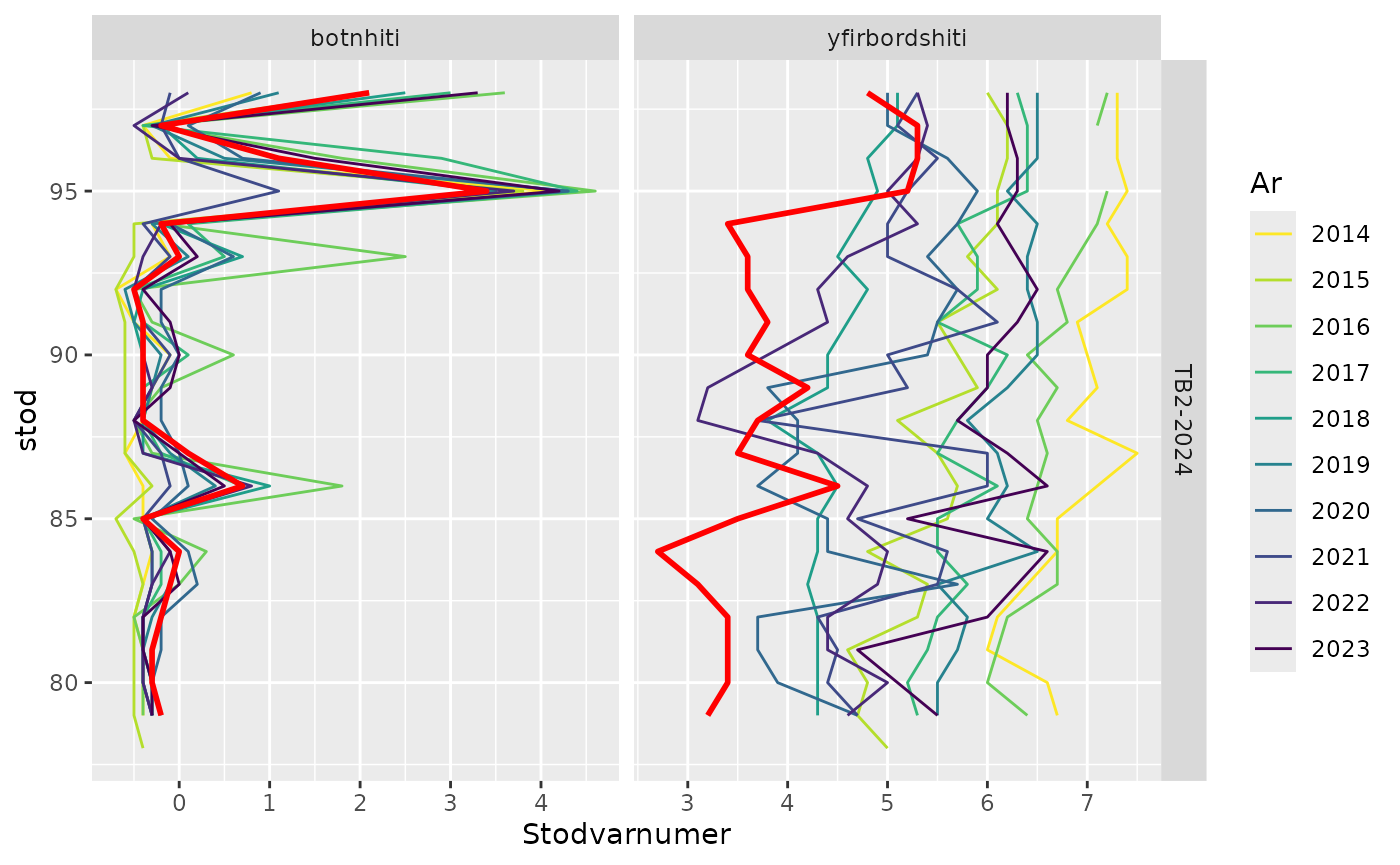
Gear metrics - time trends
res$timetrend |>
filter(leidangur %in% "TB2-2024",
var %in% c("larett_opnun", "lodrett_opnun", "vir_uti")) |>
sm_plot_violin()
#> Warning: Removed 943 rows containing non-finite outside the scale range
#> (`stat_ydensity()`).
#> Warning: Removed 943 rows containing missing values or values outside the scale range
#> (`geom_point()`).
#> Warning: Removed 5 rows containing missing values or values outside the scale range
#> (`geom_line()`).
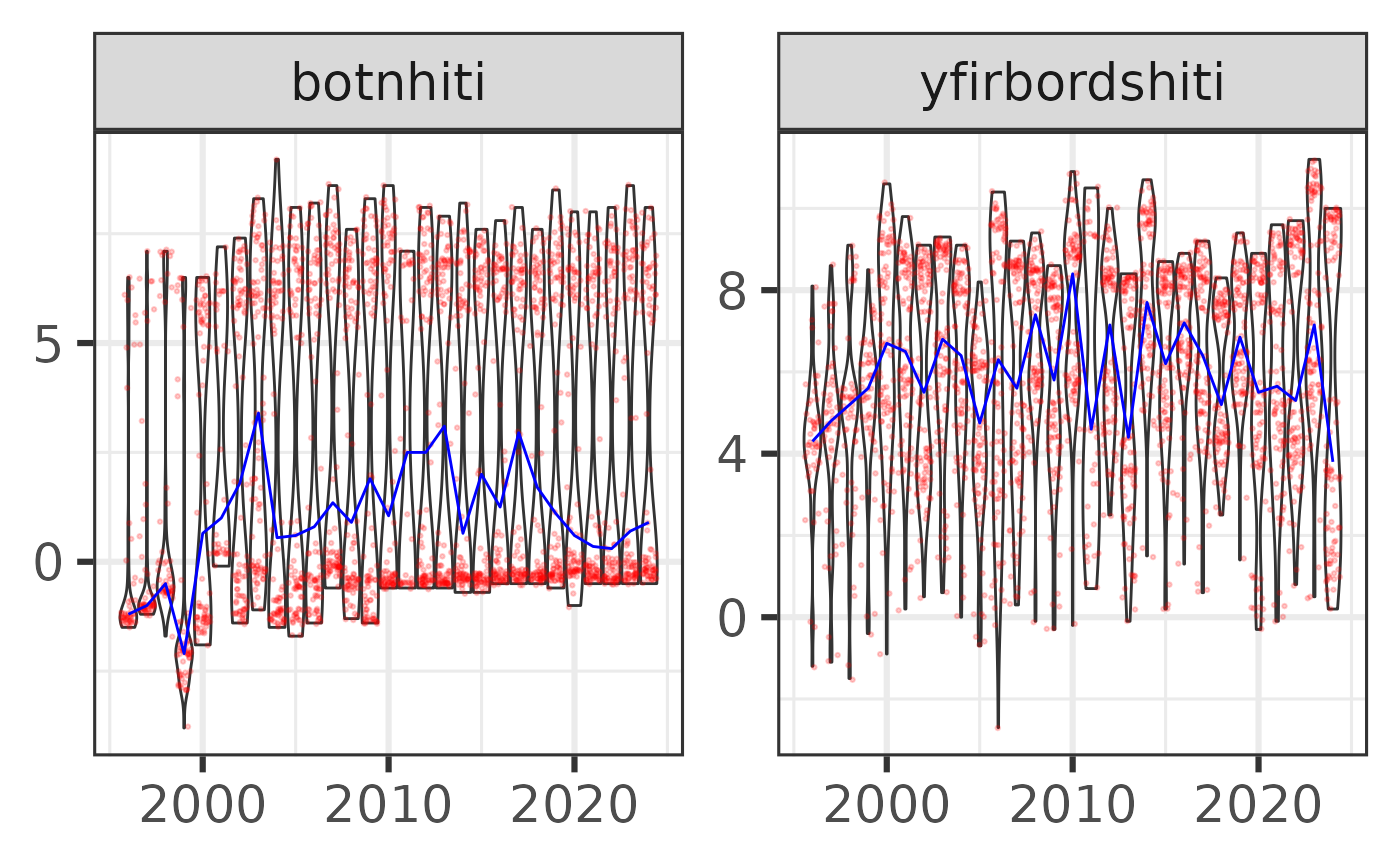
Temperature - time trends
res$timetrend |>
filter(leidangur %in% "TB2-2024",
var %in% c("botnhiti", "yfirbordshiti")) |>
sm_plot_violin()
#> Warning: Removed 115 rows containing non-finite outside the scale range
#> (`stat_ydensity()`).
#> Warning: Removed 115 rows containing missing values or values outside the scale range
#> (`geom_point()`).