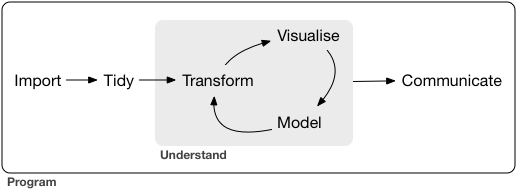
Import and export
Needed library:
library(tidyverse)
library(janitor)R is not a database software so one normally has to import the data from some other sources.
Entering data directly into R
Although R is not a good data entry medium it is possible. E.g. to one can create vectors by:
weight <- c( 1, 5, 3, 2, 6)
length <- c(10, 17, 14, 12, 18)Or one could generate a data frame by:
d <-
tibble(weight = c( 1, 5, 3, 2, 6),
length = c(10, 17, 14, 12, 18))Or one could edit a data frame using the {editData}-package. But that said R is not a good data entry medium and normally only done on an ad hoc basis.
Importing data from plain text files
A lot of functions in R deal with reading in text files in various formats. We have already used the read_csv-function.
w <- read_csv(file = "ftp://ftp.hafro.is/pub/data/csv/minke.csv")Text files (like above) that reside on the web can normally be read in directly into R. However some other files like Excel can not. If we know the url-path of any files we can first download them before importing them. Here we will just use the minke-file as an introductory example, saving the file in the directory “data-raw” in our project directory using the download.file-function:
if(!dir.exists("data-raw")) dir.create("data-raw") # create if not exists
download.file(url = "ftp://ftp.hafro.is/pub/data/csv/minke.csv",
mode = "wb",
destfile = "data-raw/minke.csv")We can then simply read the file from our computer:
w <- read_csv("data-raw/minke.csv")read_csv # US style csv file (column separator is ',' and dec '.'
read_csv2 # Alternative style csv file (column separator is ';' and dec ','
read_tsv # Tab deliminated data, US style decimal (.)
read_tsv2 # Tab deliminated data, style decimal (,)
read_delim # The generic read function
# Base R functions
read.fortran # Fotran formated text
readLines # Raw lines from the fileOn file paths
The first argument in read_xxx functions is the file name, including the path. If the file is in the current working directory (use getwd() to get information of you current working directory) one can simply write the following command:
w <- read_csv("minke.csv")If the data file are in folder called “data-raw” within the working directory:
w <- read_csv('data-raw/minke.csv')One can also use absolute paths like:
read_csv("~/edu/crfmr/data-raw/minke.csv")
read_csv("C:/Users/username/Documents/edu/crfmr/data-raw/minke.csv")Absolute paths are however specific to each computer directory structure and it is thus not recommended to use if you are distributing the project to other collaborators.
If the data is not in the current working directory tree one may use:
read_csv("../***.csv") # One folder up
read_csv("../../***.csv") # Two folders up
read_csv("../../data-raw/***.csv") # Two folders up, one folder downAgain, if sharing a project this may not be reproducable.
Sanity checks
After the data has been imported one frequently checks the data to see what was imported is as expected. Some common functions are:
head(d) # shows the top 6 lines of d
tail(d) # shows the last 6 lines of d
dim(d) # shows the num row and col of d
names(d) # gives the column names of d
summary(d) # quick summary statistics for the columns of d
str(d) # show the structure of the data, e.g. variable types
glimpse(d) # dplyr equivalent of str that works on data framesTake particular note that the class of each variable is as you expect. E.g. in the minke case one expects that the class for length is numeric (dbl) and class for date is a date-class (dttm), not e.g. character.
Arguments
Lets generate a short inline csv-file:
tmp <-
"metadata 1: this data was collected on R/V Bjarni
metatdata 2: this research was funded by EU
ID,Date,Fishing area
1,2021-01-01,North
2,2021-01-02,North
NA,NA,NA
3,2021-02-28,South
4,2021-02-29,N/A
5,-9999,South"As is very common with datafiles this file has:
- Metadata: Here the first two lines
- Missing categorical data represented as “N/A”
- Missing date information represented as -9999
Reading the tmp “csv”-file with just the default arguements gives us this:
read_csv(tmp)# A tibble: 8 × 1
`metadata 1: this data was collected on R/V Bjarni`
<chr>
1 metatdata 2: this research was funded by EU
2 ID,Date,Fishing area
3 1,2021-01-01,North
4 2,2021-01-02,North
5 NA,NA,NA
6 3,2021-02-28,South
7 4,2021-02-29,N/A
8 5,-9999,South In order to get the data properly into R we need to overwrite some of the defaults:
read_csv(tmp,
skip = 2, # skip the first two lines
na = c("N/A", "-9999")) # Representation of missing values# A tibble: 6 × 3
ID Date `Fishing area`
<chr> <chr> <chr>
1 1 2021-01-01 North
2 2 2021-01-02 North
3 NA NA NA
4 3 2021-02-28 South
5 4 2021-02-29 <NA>
6 5 <NA> South Now in the above code we manage to get things roughly right. But take note that:
- ID and date is a character, but we kind of expected a numerical and date respectively
- We kind of expected that obervations in line 3 would be interpreted as missing
- Try to fix the na-argument in order to account for “N/A”
- Check the help for
read_csvand see if you add an arguement to skip the empty row (row 3)
read_csv(tmp,
skip = 2, # skip the first two lines
skip_empty_rows = TRUE,
na = c("NA", "N/A", "-9999")) # Representation of missing valuesBut what happened to the date in ID 4??
Variable names
The variables names in imported files are often lengthy, contain combination of upper and lower cases and often have spaces. This mean downstream coding is often cumbersome though doable. To make coding less so, it is strongly adviced that you:
- Keep variable names as short as possible
- Use only lower case letters
- use “_” instead of a space in the name
janitor::clean_names does the last two parts pretty well and dplyr::rename the first.
d <- read_csv(tmp,
skip = 2,
na = c("NA", "N/A", "-9999")) |>
clean_names() |>
rename(area = fishing_area)
d# A tibble: 6 × 3
id date area
<dbl> <date> <chr>
1 1 2021-01-01 North
2 2 2021-01-02 North
3 NA NA <NA>
4 3 2021-02-28 South
5 4 NA <NA>
6 5 NA South- Read the help file on `remove_empty`` and apply it to the above dataframe such that row 3, which has no data if dropped.
- Check the difference with what you get when using
drop_nain the pipeflow instead.
d |>
remove_empty(which = "rows")
d |>
drop_na()Writing files
Writing a csv-file is as simple as:
write_csv(w, "data-raw/my-minke.csv")write_rds() store data in R’s custom binary format called RDS preserving the exact object format.
write_rds(w, "data-raw/my-minke.rds")These can be read in using read_rds():
read_rds("data-raw/my-minke.rds")# A tibble: 190 × 13
id date lon lat area length weight age sex
<dbl> <dttm> <dbl> <dbl> <chr> <dbl> <dbl> <dbl> <chr>
1 1 2004-06-10 22:00:00 -21.4 65.7 North 780 NA 11.3 Female
2 690 2004-06-15 17:00:00 -21.4 65.7 North 793 NA NA Female
3 926 2004-06-22 01:00:00 -19.8 66.5 North 858 NA 15.5 Female
4 1333 2003-09-30 16:00:00 -21.6 65.7 North 567 NA 7.2 Male
5 1334 2003-09-25 15:00:00 -15.6 66.3 North 774 NA 12.3 Female
6 1335 2003-09-16 16:00:00 -18.7 66.2 North 526 NA 9.6 Female
7 1336 2003-09-12 17:00:00 -21.5 65.7 North 809 NA 17.3 Female
8 1338 2003-09-09 12:00:00 -22.8 66.1 North 820 NA 13.8 Female
9 1339 2003-08-31 13:00:00 -17.5 66.6 North 697 NA 12.2 Male
10 1341 2003-08-30 17:00:00 -14.7 66.2 North 777 NA 15.4 Male
# ℹ 180 more rows
# ℹ 4 more variables: maturity <chr>, stomach.volume <dbl>,
# stomach.weight <dbl>, year <dbl>- Write the “d” table as rds
- Read the table in again
d |> write_rds("data-raw/tmp.rds")
read_rds("data-raw/tmp.rds")But what happened to the date in ID 4??
Importing data from excel sheets
The readxl-package provides “light-weight” support to read in Excel files directly into R. The minke data is avalaible in an excel format called minke.xlsx. You can either download it onto your computer via the usual point and mouse click or use the download.file function:
download.file(url = "https://heima.hafro.is/~einarhj/data/minke.xlsx",
destfile = "data-raw/minke.xlsx",
mode = "wb")
library(readxl)
d <-
read_excel("data-raw/minke.xlsx")
glimpse(d)Rows: 190
Columns: 12
$ id <dbl> 1, 690, 926, 1333, 1334, 1335, 1336, 1338, 1339, 1341, …
$ lon <dbl> -21.42350, -21.39183, -19.81333, -21.57500, -15.61167, …
$ lat <dbl> 65.66183, 65.65350, 66.51167, 65.67333, 66.29000, 66.17…
$ area <chr> "North", "North", "North", "North", "North", "North", "…
$ length <dbl> 780, 793, 858, 567, 774, 526, 809, 820, 697, 777, 739, …
$ weight <chr> "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "NA", "…
$ age <chr> "11.3", "NA", "15.5", "7.2", "12.3", "9.6", "17.3", "13…
$ sex <chr> "Female", "Female", "Female", "Male", "Female", "Female…
$ maturity <chr> "pregnant", "pregnant", "pregnant", "immature", "immatu…
$ stomach.volume <chr> "58", "90", "24", "25", "85", "18", "200", "111", "8", …
$ stomach.weight <chr> "31.9", "36.29", "9.42", "3.64", "5.51", "1.167", "99",…
$ year <dbl> 2004, 2004, 2004, 2003, 2003, 2003, 2003, 2003, 2003, 2…The read_excel function will by default read in the 1st data-sheet (checkout args(read_excel). To get information on what sheets are in an excel file one can use the excel_sheets function:
excel_sheets("data-raw/minke.xlsx")[1] "maesurements"If NAs are represented by something other than blank cells, set the na argument by e.g. if -9999 represents missing data then:
read_excel("data-raw/minke.xlsx", na = "-9999")One can read in a certain cell range by:
read_excel("data-raw/minke.xlsx", range = "A1:C10")# A tibble: 9 × 3
id lon lat
<dbl> <dbl> <dbl>
1 1 -21.4 65.7
2 690 -21.4 65.7
3 926 -19.8 66.5
4 1333 -21.6 65.7
5 1334 -15.6 66.3
6 1335 -18.7 66.2
7 1336 -21.5 65.7
8 1338 -22.8 66.1
9 1339 -17.5 66.6Sanity check on the object read in from Excel is an absolute must because the data can be notoriously corrupt because the user is free to do whatever in that framework either intentionally or by accident.
Other software connections
Package haven provides functions for reading in SPSS, STATA and SAS files:
library(haven)
read_sas("path/to/file") ## SAS files
read_por("path/to/file") ## SPSS portable files
read_sav("path/to/file") ## SPSS data files
read_dta("path/to/file") ## Stata filesSimilarly in the R.matlab package there is a function that reads in matlab type of data:
library(R.matlab)
readMat('path/to/file') ## Matlab data filesImporting directly from tip-files
We can read tip-files directly into R using the function foreign::read.dbf. I you have not installed the {foreign}-package before you have to install it first by running:
install.packages("foreign")Then do:
library(tidyverse)
library(foreign)If you have a raw tip-file (the suffix is .DBF) you could try something like this:
read.dbf("your/path/to/the/data/tip11.DBF", as.is = TRUE)Of course you need to replace “your/path/to/the/data/tip11.DBF” with your path of your data on your own computer. If you do not have tip file, but want to give it a go run this code:
# download some species lookup-TIP data taken from some anonymous country
download.file(url = "https://heima.hafro.is/~einarhj/data/FISHCODE.DBF", destfile = "FISHCODE.DBF", mode = "wb")
fishcode <- read.dbf("FISHCODE.DBF", as.is = TRUE)
glimpse(fishcode)Rows: 797
Columns: 10
$ NODC_COD <chr> "8741000000", "8835000000", "8861010206", "9202000000", "88…
$ SC_NAME <chr> "O+:ELOP. O:ANG. O-:ANG.", "O+:ACAN. O:PER. O-:PER.", "SPHO…
$ CMN_NAME <chr> NA, NA, NA, NA, "AMBERJACK,GREATER", "AMBERJACK,LESSER", "A…
$ ALP_CODE <chr> NA, NA, NA, NA, "GAM", "LAM", NA, NA, NA, NA, NA, "FAN", NA…
$ AREA <chr> "( )", "( )", NA, NA, "(11)", "(11)", NA, NA, NA, NA, NA,…
$ NMFS_COD <chr> "NONE", "NONE", "NONE", "NONE", "1812", "1815", "60", "NONE…
$ FISHERY <chr> NA, NA, NA, NA, "RF", "RF", "EG", "EG", "EG", "EG", "RF", "…
$ PR_CODE <chr> NA, NA, "741", NA, "112", NA, "42", NA, "44", "43", "578", …
$ OECS_CODE <chr> NA, NA, NA, NA, "CARADU", NA, "ENGR", "ENGRLY", "ENGRHE", "…
$ LOCAL_NAME <chr> NA, NA, NA, NA, "AMBERJACK,GREATER", "AMBERJACK,LESSER", "A…